Scroll to:
Polymorphisms in TLR4 gene associated with risks of bovine mastitis development
https://doi.org/10.29326/2304-196X-2025-14-1-76-81
Abstract
Introduction. Inflammatory mammary diseases in cows remain the most common challenge in dairy industry, notwithstanding the improved preventive measures and treatment schemes. One of the methods to prevent mastitis in dairy cows is the genetic selection of the most disease-resistant individuals. Toll-like receptor 4 (TLR4) plays a central role in the innate immune response. There are publications about TLR4 significance for mastitis development, its genetic polymorphisms associated with somatic cell counts.
Objective. Determination of genetic diversity and association with the development of clinical mastitis for three polymorphic locuses of TLR4.
Materials and methods. To achieve the objective cattle health history (n = 421) was used, subclinical mastitis was diagnosed using rapid test for somatic cell counting in milk, TaqMan real-time polymerase chain reaction was used for genotyping of cattle for rs8193046, rs8193060, rs29017188 polymorphisms. Results. Association studies established that rs8193046 и rs29017188 polymorphisms are the most promising candidates to be used in selection programs aimed at mastitis risk mitigation in the Ural populations. For rs8193060 no reliable results of association tests are obtained, though risk of mastitis in GCG haplotype-animals (for SNP rs8193046, rs8193060, rs29017188 alleles) is statistically lower.
Conclusion. It is noted that the abovementioned polymorphisms can be used for marker-assisted selection of cattle to prevent risks of mastitis in the populations in the Ural.
For citations:
Bytov M.V., Osipova Yu.A., Yusupova Ch.R., Zubareva V.D. Polymorphisms in TLR4 gene associated with risks of bovine mastitis development. Veterinary Science Today. 2025;14(1):76-81. https://doi.org/10.29326/2304-196X-2025-14-1-76-81
INTRODUCTION
Inflammation of a mammary gland (mastitis) is one of the costliest diseases in cattle. Preventive measures and treatment regimens are being optimized, including in order to save labor and expenses. Such measures include preventive vaccination and administration of various antimicrobial drugs [1][2][3]. An alternative to reduce mastitis incidence in farms is genetic selection. Long-term selection of dairy cattle for high milk flow, preferred due to machine milking, resulted in weakening of the mammary streak canal sphincter that represents a physical barrier for pathogen entry [4]. Susceptibility to mastitis is based on a number of factors, both external (nutrition, keeping practices, stress factors, milking techniques) and internal (immune mechanisms, important to be understood in order to increase the resistance of animals) [5].
The immune response plays a key role in the disease pathogenesis. Toll-like receptor 4 (TLR4), as an intrinsic immune receptor, exhibits widespread in vivo expression and its dysregulation significantly contributes to the onset of various diseases, encompassing cardiovascular disorders, neoplastic conditions, and inflammatory ailments [6]. The search for associations with colibacillosis risks revealed that TLR4 (rs8193046) gene polymorphism G* allele frequency was higher in diarrheic calves than in control animals [7]. In a study of the association of single nucleotide polymorphism (SNP) with the risk of paratuberculosis caused by Mycobacterium avium, it was shown that A/G heterozygotes produced a higher risk of this infectious disease [8]. An experiment aimed to find polymorphism haplotypes in TLR4 gene and conducted in different cattle populations revealed that the A* allele is present in all haplotypes and might negatively effect on мilk somatic cells. The C* allele also has a negative effect on this value and the G* allele might positively effect on milk somatic cells [9]. It is worth noting that in this study, no corrections for multiple comparisons were made in the search for statistically significant haplotypes. At the same time, in a study of the some SNP associations with risks of subclinical mastitis, it was shown that individuals with G/G genotype had higher average somatic cell counts [10].
In the case of rs8193060, indications were obtained for the association with reproductive traits: incidence of cystic ovaries, early reproductive disorders, calving ease, and production longevity [11]. There is evidence of the genetic association of polymorphisms with paratuberculosis infection, moreover it was established that the C/T genotype might be beneficial [12]. The study aimed to find polymorphism haplotypes in the TLR4 gene for rs8193060 produced ambiguous results: the C* allele might confer both positive and negative effects [9]. The analysis of rs8193060 associations with somatic cell counts showed that T/T genotype is not beneficial [10].
The rs29017188 polymorphism has the highest pleiotropic effect based on full-genome studies. There is evidence of its effect on the calving interval [13], lactation persistence [14] and milk composition [15].
Unfortunately, to date, no mechanisms have been identified for how exactly TLR4 gene polymorphisms affect the body’s immune functions. Bhat R. R. et al. described the supposed mechanism of SNPs falling in TLR4 promoter and 5’ untranslated region. Researchers have found absence of heterozygous condition in these loci in individuals with susceptibility to mastitis, which is most likely due to transcriptional factor binding profile, which ultimately changes the expression of this gene [16].
Based on the above, the aim of the study was to analyze the genetic diversity and to search for associations of TRL4 polymorphisms with the risks of mastitis in cattle.
MATERIALS AND METHODS
For genotyping of 421 cattle for rs8193046, rs8193060, rs29017188, the protocol previously described by A. Q. de Mesquita et al. [10] with a change in oligonucleotides for rs8193046 was used (Table 1). Herewith all animals were genotyped for rs8193060 and rs29017188, and 387 out of them for rs8193046. Animals from five farms of the Ural region were used.
Table 1
Oligonucleotide sequences
|
SNP |
Oligonucleotide sequence |
Amplicon length, bp |
|
rs8193046 |
F, GAGAGGAGAGTTGCTTGGAAGTCT |
107 |
|
R,GCTCCATGCACTGGTAACTAATGT |
||
|
P1, [HEX]CAGGAAGACACCGCA[ BHQ1] |
||
|
P2, [ROX]CAGGAAGACACCACA[ BHQ2] |
||
|
rs8193060 |
F, CCACTCGCTCCGGATCCT |
79 |
|
R,CCTTGGCAAATTCTGTAGTTCTTG |
||
|
P1, [HEX]ACTGCAGTTTCAACCGTATC[ BHQ1] |
||
|
P2, [ROX]ACTGCAGCTTCAACCGTA[ BHQ2] |
||
|
rs29017188 |
F, CCAGCTTCCTCTTGTTGTTACTTCA |
150 |
|
R,CGGGAGGAGAGGAAGTGAGA |
||
|
P1, [HEX]TATTTATCTCCTCTGCCACCGGA[ BHQ1] |
||
|
P2, [ROX]TTATCTCCTCTGCCACCCGAG[ BHQ2] |
The criterion for inclusion an animal into a risk group was clinical and subclinical mastitis in the disease history. Rapid tests measuring somatic cell counts were used to diagnose subclinical mastitis. If no mastitis had been recorded in the animal during three lactation periods, and the rapid test showed a negative result, the animal was considered resistant to mastitis. Blood was collected from the tail vein of all animals into vacutainer tubes containing EDTA (ethylenediaminetetraacetic acid) as an anticoagulant.
The genotype distribution was analyzed for compliance with Hardy – Weinberg principle; linkage disequilibrium and the Shannon diversity index were calculated using GenAlEx package (version 6.5) for Microsoft Excel [17]. Linkage disequilibrium graphs were constructed using SRplot web tool [18].
Association tests for each SNP individually, the search for the most common haplotypes and their associations with mastitis risks were performed by SNPassoc R package (version 2.1.0) [19].
RESULTS AND DISCUSSION
The allelic diversity and genotype distribution are shown in Table 2. When calculating the Hardy – Weinberg proportion, 1 degree of freedom was used. Based on the analysis results, a statistically significant deviation from equilibrium allele distribution was revealed for rs8193046 polymorphism. Such deviations can occur for a number of reasons: selective pressure, genotyping errors, inbreeding. The most likely explanation is the pressure of artificial selection.
Table 2
Genotype distribution, allele frequency, and p-value of Hardy – Weinberg equilibrium
|
Locus |
Genotype |
Number of animals |
Genotypic frequency, % |
Alleles |
Allelic frequency |
X² (p-value) |
|
rs8193046 G > A |
A/A A/G G/G |
89 145 153 |
23.0 37.5 39.5 |
A* G* |
323 451 |
20.397 (< 0.0001) |
|
rs8193060 T > C |
C/C C/T T/T |
184 191 46 |
43.7 45.4 10.9 |
C* T* |
559 283 |
0.116 (0.734) |
|
rs29017188 G > C |
C/C C/G G/G |
75 212 134 |
17.8 50.4 31.8 |
C* G* |
362 480 |
0.314 (0.575) |
Based on the linkage disequilibrium analysis, it can be concluded that the pairs of rs8193046 and rs8193060 alleles, as well as rs29017188 and rs8193060 demonstrate linkage disequilibrium: R² = 0.2 and R² = 0.4, respectively (Fig. 1).
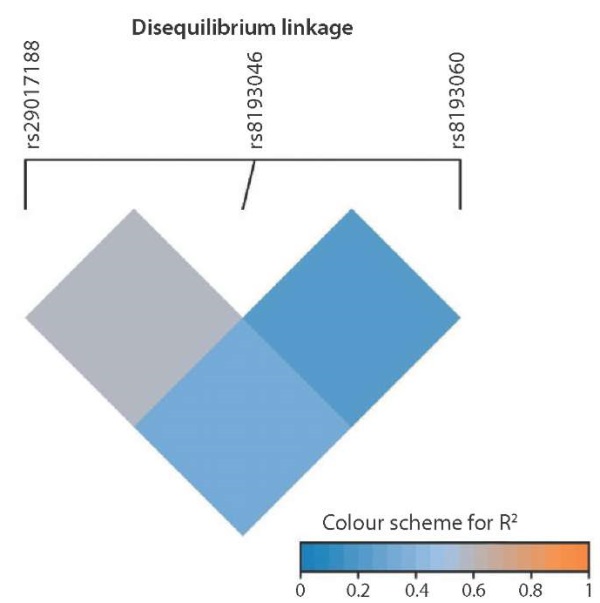
Fig. 1. Linkage disequilibrium (R²) plot of the studied SNPs
The genetic diversity of the studied cattle populations was also evaluated by TLR4 gene polymorphisms. The Shannon diversity index approaching 1 reflects a high diversity level. Thanks to the analysis, it was found that the diversity between populations is low (D’ = 0.015). However, on average, a higher diversity index value (D’ = 0.403) can be observed within populations, from which it can be concluded that they are genetically stable (Fig. 2). Since there are no differences in the diversity for these polymorphisms between the studied populations, further tests were performed jointly.
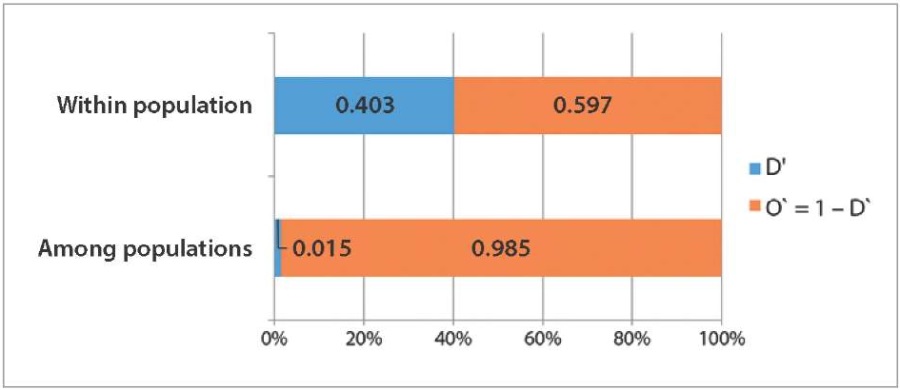
Fig. 2. Shannon diversity index for intra- and interpopulation assessment: D’ – diversity index; O’ – overlap index
Association tests were performed individually for each of the TLR4 gene polymorphisms to identify associations with mastitis risks. А summary of the results is presented in Table 3: rs29017188 SNP showed the largest number of statistically significant inheritance models, including taking into account the Bonferroni correction, while the recessive inheritance of mastitis risk was significant for rs8193046 and rs29017188. The recessive inheritance for rs8193046 also has the lowest Akaike information criterion (AIC) value, and the odds ratio (OR) suggests that animals with A/A genotype have a higher risk of mastitis. For rs29017188, the recessive inheritance model also had the lowest AIC value, OR for the C/C genotype was 2.30, this means the risk of mastitis is presumably more than 2 times higher in individuals of the studied populations.
Table 3
Results of association tests for each of the SNPs for five inheritance models
|
SNP |
Codominant |
Dominant |
Recessive |
Overdominant |
log-additive |
|
rs8193046 |
0.04887* |
0.17175 |
0.01478* |
0.47568 |
0.02935* |
|
rs8193060 |
0.51985 |
0.34612 |
0.73568 |
0.25287 |
0.58147 |
|
rs29017188 |
0.00671* |
0.06545 |
0.00248* |
0.57736 |
0.00368* |
|
* p-value ≤ 0.05; in bold – p-value ≤ 0.016 (Bonferroni correction). |
|||||
The results of the haplotype search showed that ACC, GCG, and GTG (for the rs8193046, rs8193060, and rs29017188 SNP alleles, respectively) are the most common, accounting for more than 85% of the total sample (Table 4).
Table 4
Haplotypes and their associations with mastitis risks
|
rs8193046, rs8193060, rs29017188 |
Frequency |
OR |
95% confidence interval |
p-value |
|
ACC |
0.3491 |
1.00 |
reference haplotype |
– |
|
GCC |
0.0643 |
0.86 |
0.45–1.64 |
0.6515 |
|
GCG |
0.2145 |
0.53 |
0.36–0.80 |
0.0022* |
|
GTG |
0.3020 |
0.74 |
0.52–1.05 |
0.0894 |
|
Other rare haplotypes |
0.0701 |
0.60 |
0.33–1.12 |
0.1083 |
|
* p-value ≤ 0.05; in bold – p-value ≤ 0.016 (Bonferroni correction). |
||||
Animals with the GCG haplotype have a statistically significant lower risk of mastitis (for SNPs rs8193046, rs8193060, rs29017188). We assume that an increase in the proportion of individuals with this haplotype in farms where mastitis in dairy cows is challenging may have a positive effect on the disease occurrence. The results of the search for associations of haplotypes with phenotypes coincide with the results of the identification of individual polymorphism associations and are more consistent with the data obtained by P. Wang et al. [9].
CONCLUSION
In the course of the study, the genetic diversity of the Ural dairy cattle populations was analyzed. The rs8193046 polymorphism of the TRL4 gene revealed deviation from the Hardy – Weinberg equilibrium, which is most likely due to the influence of artificial selection pressure.
Based on the results of association tests, it was assumed that SNP rs8193046 and rs29017188 are the most promising candidates for use in breeding programs to reduce the risk of mastitis in the studied populations. It is worth noting the low effectiveness of genomic estimate extrapolation even among populations of the same breed; however, the results obtained during the study coincide with the previously published data [9]. The GCG haplotype for rs8193046, rs8193060, and rs29017188 was found to be statistically significant based on the association tests. This haplotype can be probably used for positive selection to reduce the risk of clinical mastitis in dairy cattle populations.
References
1. Isakova M. N., Ryaposova M. V., Oparina O. Yu. Changesin the indices of general resistance of the organism of cows on the background of the use of anti-mastitis vaccines. Bulletin of Veterinary Pharmacology. 2019; (1): 91–95. https://doi.org/10.17238/issn2541-8203.2019.1.91 (in Russ.)
2. Isakova M. N., LysovaYa. Yu. The effect of the nisin-based pharmaceutical formulation used in the treatment plan for cows with subclinical mastitis on the milk microbiota. Veterinary Science Today. 2024; 13 (3): 261–268. https://doi.org/10.29326/2304-196X-2024-13-3-261-268
3. Drozdova L. I., Barkova A. S., Isakova M. N., Larionov L. P., Permikin V. V., Starikov N. M., Khonina T. G. Evaluating wound-healing effect of silicon-zinc-boron-containing glycerohydrogel and its effect on mammary glands of high producing dairy cows. Veterinary Science Today. 2023; 12 (4): 322–330. https://doi.org/10.29326/2304-196X-2023-12-4-322-330
4. Brajnik Z., Ogorevc J. Candidate genesfor mastitis resistance in dairy cattle: a data integration approach. Journal of Animal Science and Biotechnology. 2023; 14:10. https://doi.org/10.1186/s40104-022-00821-0
5. Zemanova M., Langova L., Novotná I., Dvorakova P., Vrtkova I., Havlicek Z. Immune mechanisms, resistance genes, and their roles in the prevention of mastitisin dairy cows. Archives Animal Breeding. 2022; 65 (4): 371–384. https://doi.org/10.5194/aab-65-371-2022
6. Wei J., Zhang Y., Li H.,Wang F.,Yao S. Toll-like receptor 4: A potential therapeutic target for multiple human diseases. Biomedicine & Pharmacotherapy. 2023; 166:115338. https://doi.org/10.1016/j.biopha.2023.115338
7. Judi H., Judi R., Saqban A.-K. Molecular study of colibacillosis susceptibility in calves and lambs. Nano Biomedicine and Engineering. 2020; 12 (2): 153–159. https://doi.org/10.5101/nbe.v12i2.p153-159
8. Gopi B., Singh R. V., Kumar S., Kumar S., ChauhanA., KumarA., Singh S. V. Single-nucleotide polymorphisms in CLEC7A, CD209 and TLR4 gene and their association with susceptibility to paratuberculosis in Indian cattle. Journal of Genetics. 2020; 99:14. https://doi.org/10.1007/s12041-019-1172-4
9. Wang X. P., Luoreng Z. M., Gao S. X., Guo D. S., LiJ. Y., Gao X., et al. Haplotype analysis of TLR4 gene and its effects on milk somatic cellscore in Chinese commercial cattle. Molecular Biology Reports. 2014; 41 (4): 2345–2351. https://doi.org/10.1007/s11033-014-3088-7
10. De MesquitaA. Q., e Rezende C. S. M., de MesquitaA. J., Jardim E. A. G., Kipnis A. P. J. Association of TLR4 polymorphisms with subclinical mastitisin Brazilian holsteins. Brazilian Journal of Microbiology. 2012; 43 (2): 692–697. https://doi.org/10.1590/S1517-83822012000200034
11. Novák K., Valčíková T., Samaké K., Bjelka M. Association of variants in innate immune genes TLR4 and TLR5 with reproductive and milk production traitsin Czech Simmental cattle. Genes. 2024; 15 (1):24. https://doi.org/10.3390/genes15010024
12. Kumar S., Kumar S., Singh R. V., Chauhan A., Kumar A., Sulabh S., et al. Genetic association of polymorphisms in bovine TLR2 and TLR4 genes with Mycobacterium avium subspecies paratuberculosis infection in Indian cattle population. Veterinary Research Communications. 2019; 43 (2): 105–114. https://doi.org/10.1007/s11259-019-09750-2
13. Jecminkova K., MüllerU., Kyselova J., Sztankoova Z., Zavadilova L., Stipkova M., Majzlik I. Association of leptin, toll-like receptor 4, and chemokine receptor of interleukin 8 C-X-C motifsingle nucleotide polymorphisms with fertility traits in Czech Fleckvieh cattle. Asian-Australasian Journal of Animal Sciences. 2018; 31 (11): 1721–1728. https://doi.org/10.5713/ajas.17.0900
14. Sharma B. S., Leyva I., Schenkel F., Karrow N. A. Association of tolllike receptor 4 polymorphisms with somatic cell score and lactation persistency in Holstein bulls. Journal of Dairy Science. 2006; 89 (9): 3626–3635. https://doi.org/10.3168/jds.S0022-0302(06)72402-X
15. Wang M., Song H., Zhu X., Xing S., Zhang M., Zhang H., et al. Toll-like receptor 4 gene polymorphisms influence milk production traitsin Chinese Holstein cows. Journal of Dairy Research. 2018; 85 (4): 407–411. https://doi.org/10.1017/s0022029918000535
16. Bhat R. R., BhatN. N., ShabirA., Mir M. U. R., Ahmad S. B., Hussain I., et al. SNP analysis of TLR4 promoter and its transcriptional factor binding profile in relevance to bovine subclinical mastitis. Biochemical Genetics. 2024; 62 (5): 3605–3623. https://doi.org/10.1007/s10528-023-10578-4
17. Peakall R., Smouse P. E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006; 6 (1): 288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
18. Tang D., Chen M., Huang X., Zhang G., Zeng L., Zhang G., et al. SRplot: A free online platform for data visualization and graphing. PLoS ONE. 2023; 18 (11):e0294236. https://doi.org/10.1371/journal.pone.0294236
19. González J. R., Armengol L., Solé X., Guinó E., MercaderJ. M., Estivill X., Moreno V. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007; 23 (5): 644–645. https://doi.org/10.1093/bioinformatics/btm025
About the Authors
M. V. BytovRussian Federation
Maksim V. Bytov, Postgraduate Student, Junior Researcher, Department of Animal Genomics and Selection
112а Belinsky str., Ekaterinburg 620142
Yu. A. Osipova
Russian Federation
Yulia A. Osipova, Student, Laboratory Assistant, Department of Animal Genomics and Selection
112а Belinsky str., Ekaterinburg 620142
Ch. R. Yusupova
Russian Federation
Chulpan R. Yusupova, Dr. Sci. (Biology), Senior Researcher, Department of Animal Genomics and Selection
112а Belinsky str., Ekaterinburg 620142
V. D. Zubareva
Russian Federation
Vladlena D. Zubareva, Junior Researcher, Department of Animal Genomics and Selection
112а Belinsky str., Ekaterinburg 620142
Review
For citations:
Bytov M.V., Osipova Yu.A., Yusupova Ch.R., Zubareva V.D. Polymorphisms in TLR4 gene associated with risks of bovine mastitis development. Veterinary Science Today. 2025;14(1):76-81. https://doi.org/10.29326/2304-196X-2025-14-1-76-81



































