Scroll to:
Antimicrobial resistance of Salmonella spp. detected in animal products in 2022–2024
https://doi.org/10.29326/2304-196X-2025-14-3-310-318
Abstract
Introduction. Although antibiotics represent one of humanity’s greatest discoveries, their improper use can cause significant harm and lead to severe consequences. Objective. Testing of animal product samples followed by Salmonella spp. isolation, typing, identification and assessment of their antimicrobial resistance dynamics.
Materials and methods. The study was carried out at the Department for Microbiological Testing of the Vladimir Testing Laboratory of the Federal Centre for Animal Health. The disc diffusion test was used to determine bacteria resistance to antibiotics. The sizes of the microorganism growth inhibition zones were interpreted according to the Russian recommendations “Determination of the sensitivity of microorganisms to antimicrobial drugs” (IACMAC, version 2025-01), prepared on the basis of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations and using CLSI M100 standard. PETSAL® monovalent and polyvalent O- and H-sera (The Saint Petersburg Scientific Research Institute ofVaccines and Serums of the FMBA of Russia) were used for serological identification. Antimicrobial resistance genes (blaCTX-M, blaOXA10, blaDHA, blaDES, blaKPC, blaOXA48-like, blaNDM, blaVIM) were identified by real-time polymerase chain reaction using the RESISTOM test systems (“LITECH”Co. Ltd., Russia).
Results. Forty-two Salmonella spp. isolates were recovered from animal product samples in 2022–2024. S. Enteritidis was the most frequently isolated serovar, and Salmonella spp. were predominantly isolated from poultry meat products. The detected isolates demonstrated maximum resistance to benzylpenicillin, erythromycin, norfloxacin, and tetracycline. Most of the isolates showed multiple resistance to several antimicrobials. Increased resistance to cephalosporins, fluoroquinolones, tetracyclines, aminoglycosides, chloramphenicol/levomycetin and sulfamethoxazole/trimethoprim was demonstrated for Salmonella spp. isolates. No antimicrobial resistance genes were detected when the isolates were tested with real-time polymerase chain reaction.
Conclusion. The study demonstrated widespread antimicrobial resistance, including multiple resistance, among Salmonella spp. isolates detected in animal products in 2022–2024.
Keywords
For citations:
Akulich O.A., Shadrova N.B., Denisova G.S. Antimicrobial resistance of Salmonella spp. detected in animal products in 2022–2024. Veterinary Science Today. 2025;14(3):310-318. https://doi.org/10.29326/2304-196X-2025-14-3-310-318
INTRODUCTION
In 2024, the World Health Organization (WHO) published a list of 24 antibiotic-resistant bacterial pathogens. Listed gram-negative bacteria, including Salmonella spp., are of great concern due to their resistance to the latest generation of antimicrobials [1].
Antimicrobial resistance is a global challenge requiring a coordinated approach and actions at all levels: human health, agriculture, environment management, food production (WHO classified antimicrobial resistance among the top ten global public health threats in 2019) [2][3][4].
Incorrect and uncontrolled use of antibiotics is the main factor leading to the antimicrobial resistance development. Agricultural workers are particularly at risk given the fact that up to 50–80% of all antibiotics are used in the agriculture sector [5][6].
Considering that human, animal, plant and environmental health, including ecosystems, are closely interrelated and interdependent, the WHO, FAO (Food and Agriculture Organization of the United Nations), UNEP (United Nations Environment Programme) and WOAH (World Organization for Animal Health) have joined efforts to combat antimicrobial resistance within the framework of the One Health concept. The WHO established the Global Antimicrobial Resistance and Use Surveillance System (GLASS) providing standardized methods and in 2001 published the WHO Global Strategy for Containment of Antimicrobial Resistance. In 2022 the WHO launched the Global Genomic Surveillance Strategy for Pathogens with Pandemic and Epidemic Potential (2022–2032) [7].
The FAO has called for halting the use of antimicrobials for the infection prevention and as growth promoters in livestock and aquaculture sectors as a part of the combating antimicrobial resistance. Additionally, the FAO/WHO Codex Alimentarius Commission on Food Standards adopted strict standards on the maximum permissible limits for medicinal product residues.
Moreover, world leaders adopted a political declaration at the 79th session of the United Nations General Assembly, in which they committed to achieving specific targets to combat antimicrobial resistance.
In Russia, antimicrobial resistance is also regulated by legislation. In 2017, the National Strategy of the Russian Federation for Preventing the Spread of Antimicrobial Resistance in the Russian Federation to 2030 outlining the official policy aimed at limiting the spread of antimicrobial resistance was approved by Russian Federation Government Order No. 2045-r. In 2024, the Action Plan for the Implementation of the Strategy for the Prevention of the Spread of Antimicrobial Resistance in the Russian Federation by 2030 addressing the key areas such as regulatory measures, public awareness, systemic monitoring and other aspects related to antimicrobial resistance for 2025–2030 was approved by Russian Government Order No. 2214-r.
Therewith, list of veterinary medicinal products restricted for therapeutic use (approved by Order No. 771 of the Ministry of Agriculture of the Russian Federation of 18 November 2021) has been put in effect since 2022, and procedure for prescribing veterinary medicinal products and the list of prescription veterinary medicinal products (approved by Order No. 776 of the Russian Federation Ministry of Agriculture of 2 November 2022) have entered into force since 2025.
In 2024, the first BRICS International Conference on Antimicrobial Resistance was held in Moscow.
For the purpose of medicinal product residue control in food products, the following amendments were made to the Technical Regulations of the Customs Union by Decision No. 70 of the Council of the Eurasian Economic Commission of 23 June 2023: permissible limits for 75 medicinal products were established, as well as new requirements for providing information on used veterinary medicinal products were laid down.
Continuous monitoring for changes in the antimicrobial susceptibility of pathogens is one of the measures for combating the antimicrobial resistance spread. There is an on-line antimicrobial resistance map platform and other antimicrobial resistance services: web products designed for antimicrobial resistance surveillance for analysis of the data on antimicrobial resistance in Russia.
Salmonella spp. are one of the four main causes of diarrheal diseases in the world. Annually, diseases caused exclusively by Salmonella spp. claim the lives of more than 200,000 people worldwide [8][9].
Over the last 10 years, salmonellosis has remained a significant concern in the Russian Federation due to the existing risks of infection in intensively developing agricultural sector. Thus, according to the Federal Service for the Oversight of Consumer Protection and Welfare (Rospotrebnadzor) reports, salmonellosis incidence per 100,000 population was as follows in the Russian Federation: 24.68 cases in 2024; 21.45 cases in 2023; 17.10 cases in 2022 and 13.61 cases in 20211,2,3,4.
The most frequently detected serovars causing diseases in all countries are: S. Enteritidis, S. Typhimurium and S. Infantis. The frequency of other serovars detection depends on the region [10][11].
Currently, there is a rise in antibiotic-resistant infections, including those caused by Salmonella. At the same time, antimicrobial-resistant Salmonella pose a significant threat to the human and animal life due to their widespread prevalence and ability to contaminate water sources, among other transmission routes. Food products are the main factor in Salmonella transmission [12][13][14].
In the view of the above, the study was aimed at testing samples of animal products from three Central Russian regions (Vladimir, Kostroma and Ivanovo Oblasts), followed by Salmonella isolation, identification, typing and their assessment for antimicrobial resistance dynamics in 2022–2024.
MATERIALS AND METHODS
Tests were carried out at the Department for Microbiological Testing of the Vladimir Testing Laboratory, Federal Centre for Animal Health. Forty-two Salmonella isolates recovered from animal products in 2022–2024 were used for tests.
Reagents and nutrient media: buffered peptone water (HiMedia Laboratories Pvt Ltd., India), Rappaport – Vassiliadis magnesium medium (RVS-broth; Merck KGaA, Germany), selenite broth (Merck KGaA, Germany), tryptic soy agar (TSA; Scharlab S.L., Spain), xylose lysine deoxycholate agar (XLD-agar; State Research Center for Applied Microbiology and Biotechnology, Russia), bismuth sulphite agar (Merck KGaA, Germany), Mueller – Hinton agar (State Research Center for Applied Microbiology and Biotechnology, Russia).
Microbiological tests were performed according to GOST 31659-2012 “Food products. Method for the detection of Salmonella spp.”. The weighted portion of the product (25 g) was placed in a sterile bag containing 225 cm³ of buffered peptone water, homogenized for 1 min and then incubated at 37 °C for 18–20 hours.
The prepared cultures (1 mL) were re-inoculated into selective enrichment media: RVS-broth (10 mL) and selenite broth (10 mL) and incubated at temperature of (41.5 ± 1.0) °C and 37 °C, respectively, for 24 hours. Then, the material from each tube was re-inoculated by streaking using bacteriological loop according to GOST 26670-91 “Food products. Methods for cultivation of microorganisms” onto two selective agar media: XLD-agar and bismuth sulfite agar, and then incubated at 37 °C for (24 ± 3) hours.
To identify selected colonies demonstrating growth characteristic of Salmonella spp. and to prepare isolated colonies the materials were re-inoculated and then cultivated onto dried TSA supplemented with yeast extract at a temperature of 37 °C for (24 ± 3) hours.
The grown colonies were typed as Salmonella with API 20E biochemical tests (bioMérieux, France) and enzyme-linked immunosorbent assay using a Mini Vidas analyser (bioMérieux, France).
Serological identification. During the study, Salmonella spp. isolates recovered using slide agglutination test with PETSAL® dry diagnostic adsorbed mono- and polyvalent O- and H-sera (The Saint Petersburg Scientific Research Institute of Vaccines and Serums of the FMBA of Russia) were serologically identified. The serological variant of the strain was identified using serological formula in accordance with Kauffman – White scheme according to MG 4.2.4070-24 “Laboratory diagnosis of salmonellosis, detection of Salmonella in food products and environmental objects: methodical guidelines” (approved by the Chief Medical Officer of the Russian Federation on 27 September 2024).
Determination of antimicrobial resistance. The recovered Salmonella spp. isolates were tested for their susceptibility to antimicrobials with disc diffusion test according to MG 4.2.1890-04 “Determination of the susceptibility of microorganisms to antibacterial drugs: methodical guidelines”.
Antibiotics (paper disks produced by the Saint Petersburg Pasteur Institute, Russia): azithromycin 15 μg; amikacin 30 μg; amoxicillin 20 μg; ampicillin/sulbactam 10 μg; benzylpenicillin 10 U/6 μg; gentamicin 10 μg; doxycycline 30 μg; imipenem 10 μg; kanamycin 30 μg; levofloxacin 5 μg; meropenem 10 μg; norfloxacin 10 μg; sulfamethoxazole/trimethoprim 23.75/1.25 μg; streptomycin 10 μg; tetracycline 30 μg; chloramphenicol/levomycetin 30 μg; cefazolin 30 μg; cefotaxime 30 μg; cefuroxime 30 μg; ciprofloxacin 5 μg; erythromycin 15 μg.
Bacterial suspension (optical density – 0.5 according to the McFarland standard) prepared from day-old cultures of Salmonella spp. isolates grown on TSA was used for antibiotic resistance determination. VITEK BIOMERIEUX model DENSICHEK (France) densitometer was used for the suspension density determination.
Molten TSA not later than 15 minutes after its preparation was poured into sterile Petri dishes, (100 mm in diameter), 20 mL per Petri dish. The bacterial suspension, was inoculated by streaking onto dried Mueller – Hinton agar with sterile cotton swab, then the discs were placed onto the agar (4 discs per Petri dish). After placing antibiotic discs, the Petri dishes were incubated at 37 °C for (18 ± 2) hours. The results were assessed according to the microbial growth inhibition zones formed around the discs, measured at accuracy of 1 mm on a dark matte surface at a distance of about 30 cm from the eyes using a ruler at an angle of 45°.
The results were interpreted according to the Russian recommendations “Determination of microorganism susceptibility to antimicrobials” (IACMAC, version 2025-01) prepared on the basis of the recommendations of the European Committee for Antimicrobial Susceptibility Testing (EUCAST), and using CLSI M100 standard. The EUCAST susceptibility assessment and interpretation approaches are currently considered theoretically sound [15][16][17].
Real-time polymerase chain reaction. Sorb-GMO-B kit (Syntol, Russia) was used for the extraction of Salmonella spp. DNAs.
Salmonella spp. isolates were examined for their molecular and genetic properties as well as for antimicrobial resistance genes (blaCTX-M, blaOXA10, blaDHA, blaGES, blaKPC, blaOXA48-like, blaNDM, blaVIM) using RESISTOM test systems (Lytech, Russia) according to the manufacturer’s instruction.
RESULTS AND DISCUSSION
Forty-two Salmonella spp. isolates were detected in animal product samples: 15 isolates in 2022, 11 isolates in 2023, 16 isolates in 2024.
The morphological and cultural properties of Samonella spp. isolates were characteristic of their family and genus.
Figure 1 shows that Salmonella spp. were most often detected in poultry meat products – 36 isolates (85.7%), particularly in chicken meat – 24 isolates (57.1%). Therewith, according to the European Centre for Disease Prevention and Control (ECDC), poultry meat and poultry meat preparations are the most commonly infected with Salmonella spp. at the stage of their distribution in the European Union. Turkey meat and turkey meat products as well as pork were also found to be highly contaminated [10][18].
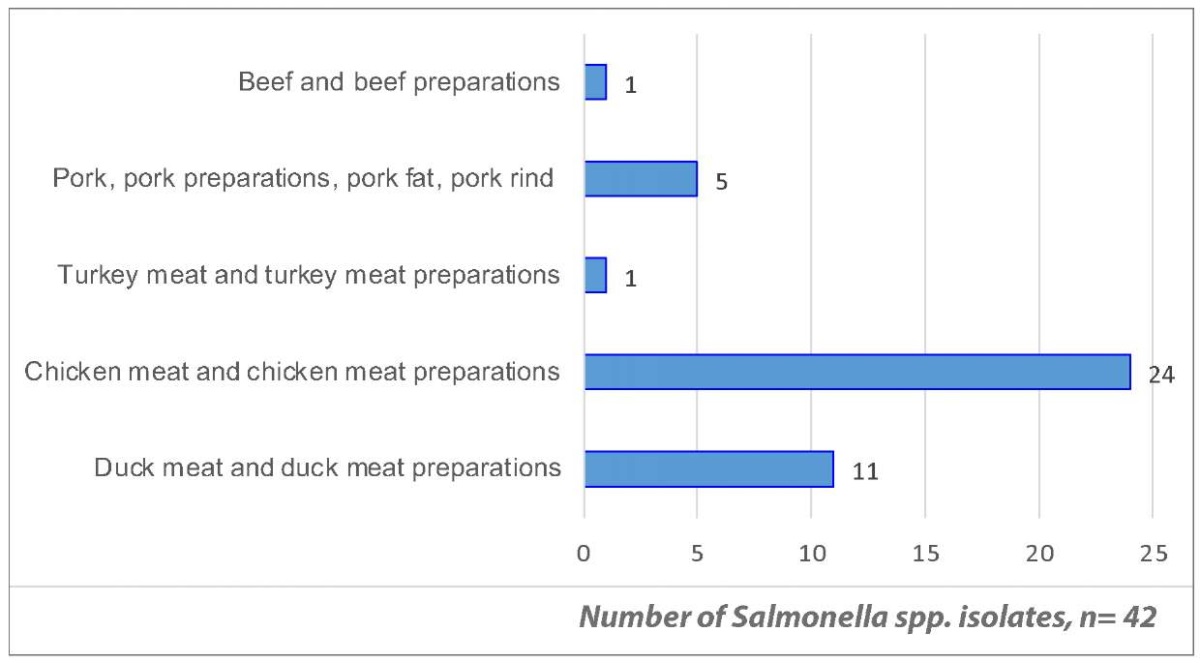
Fig. 1. Frequency of Salmonella spp. isolate detection in animal product samples in 2022–2024, by isolation source
Serological identification showed that the most Salmonella spp. isolates belonged to O:9 (D1) group – 18 isolates (42.9%) and to O:7 (C1) group – 13 isolates (30.9%), 7 isolates (16.7%) belonged to O:4 (B) group, 4 isolates (9.5%) belonged to O:8 (C2–C3) group. The results are shown in Figure 2.
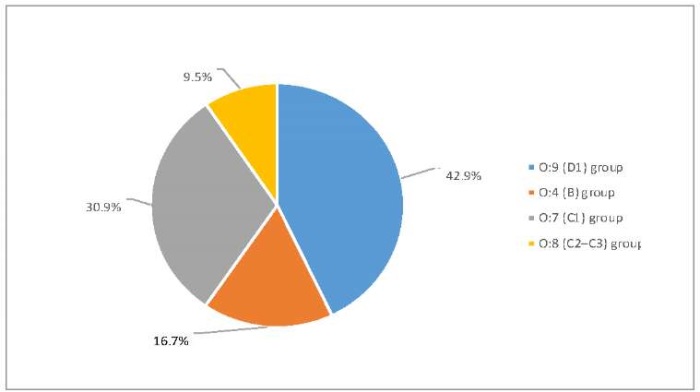
Fig. 2. Identification of O-groups of Salmonella spp. isolates recovered from samples of animal products in 2022–2024
Serotyping showed that the most often detected Salmonella spp. were as follows (Fig. 3): S. Enteritidis – 14 (33.3%), S. Blegdam – 3 (7.1%), S. Derby – 2 (4.8%). Therewith, S. Enteritidis and S. Derby were more often detected in duck products, while S. Blegdam – in chicken meat. Also, among 17 non-typeable Salmonella spp. isolates (40.5%), 11 isolates (26.2%) were classified to the O:7 (C1) group.
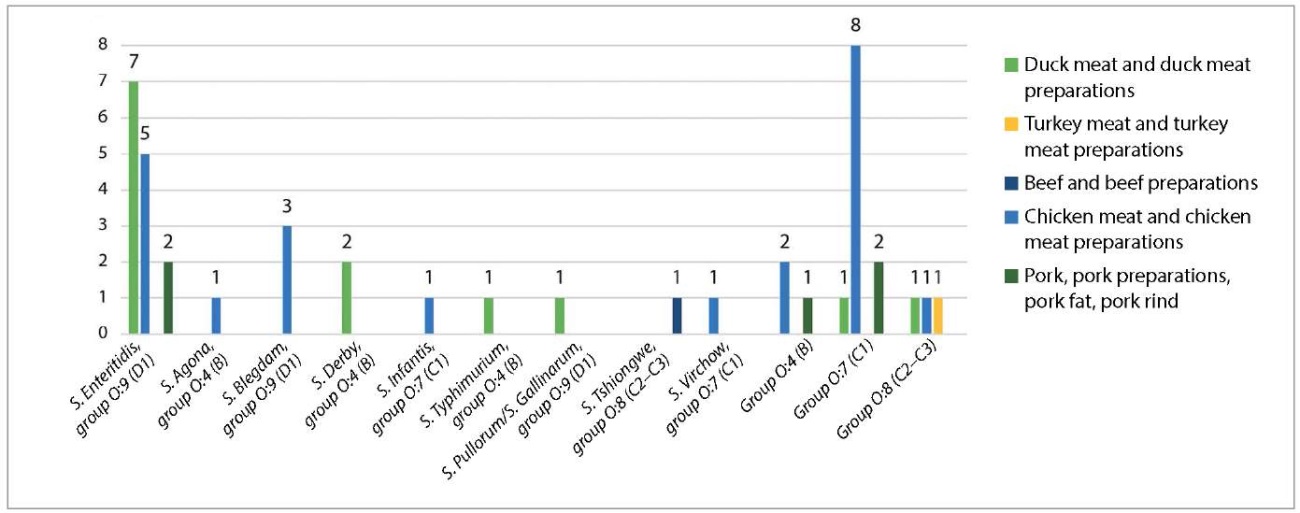
Fig. 3. Serotyping of Salmonella spp. isolates recovered from animal product samples in 2022–2024
During the study, Salmonella spp. isolates were tested for their resistance to 21 antimicrobials. The results are shown in Figure 4.
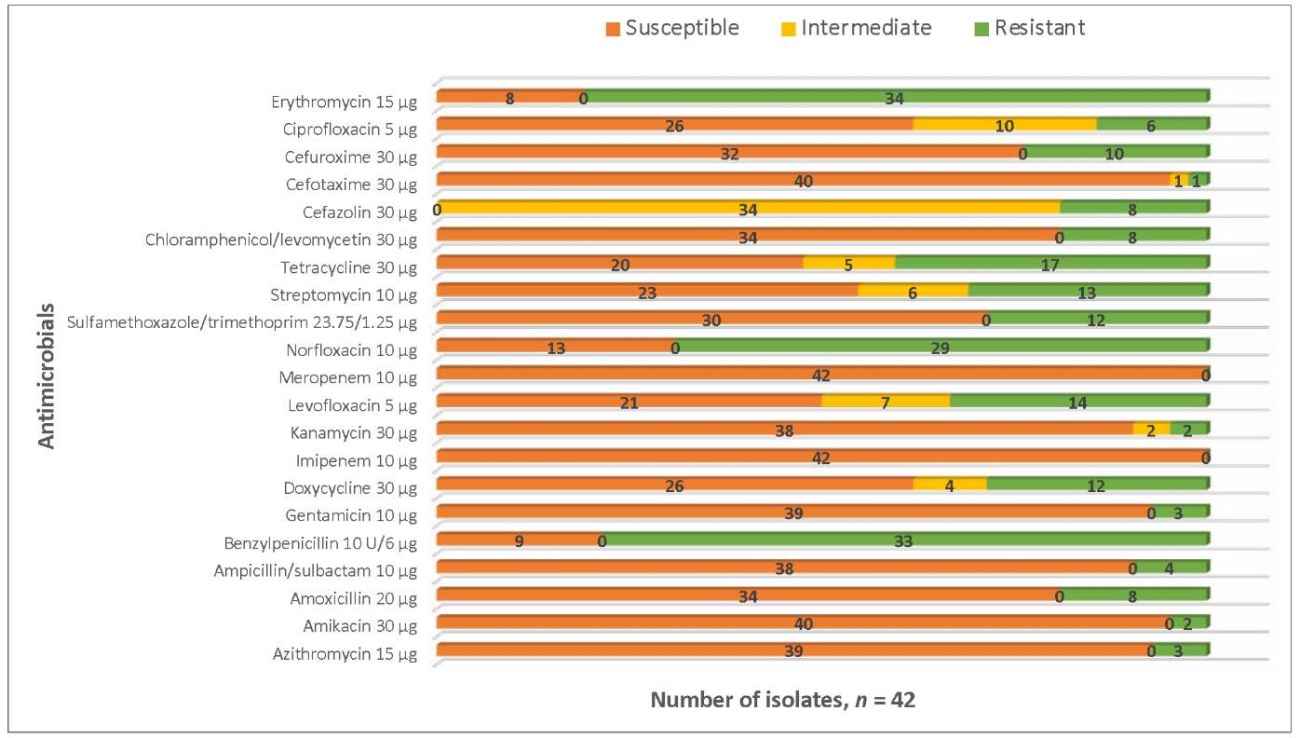
Fig. 4. Antimicrobial resistance of Salmonella spp. isolates recovered from animal products in 2022–2024
Salmonella spp. isolates recovered from animal products in 2022–2024 demonstrated relatively high level of common resistance to some antibiotics.
Examined Salmonella spp. isolates were the most frequently resistant to erythromycin (80.9%), benzylpenicillin (78.6%), norfloxacin (69.0%) and tetracycline (40.5%).
It should be noted that all Salmonella spp. isolates were susceptible to meropenem and imipenem.
In 2025, the “European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2022–2023” was published. It states that most Salmonella spp. isolates are also resistant to tetracycline and sulfonamides. Moreover, there is a trend for increase in resistance to ciprofloxacin and third generation cephalosporins in several countries [19].
These and other studies [20] indicate that Salmonella spp. susceptibility monitoring is crucial because of their increasing resistance to some critically important antimicrobials.
Also, tests showed that 90% of Salmonella spp. isolates were resistant to more than one of the tested antibiotics. In addition, 38% of the isolates were resistant to more than three classes of antimicrobials.
Figure 5 shows that 13 isolates (31.0%) demonstrated resistance to three antimicrobials, 8 isolates (19.0%) – to two antimicrobials and 5 isolates (11.9%) – to eight antimicrobials.
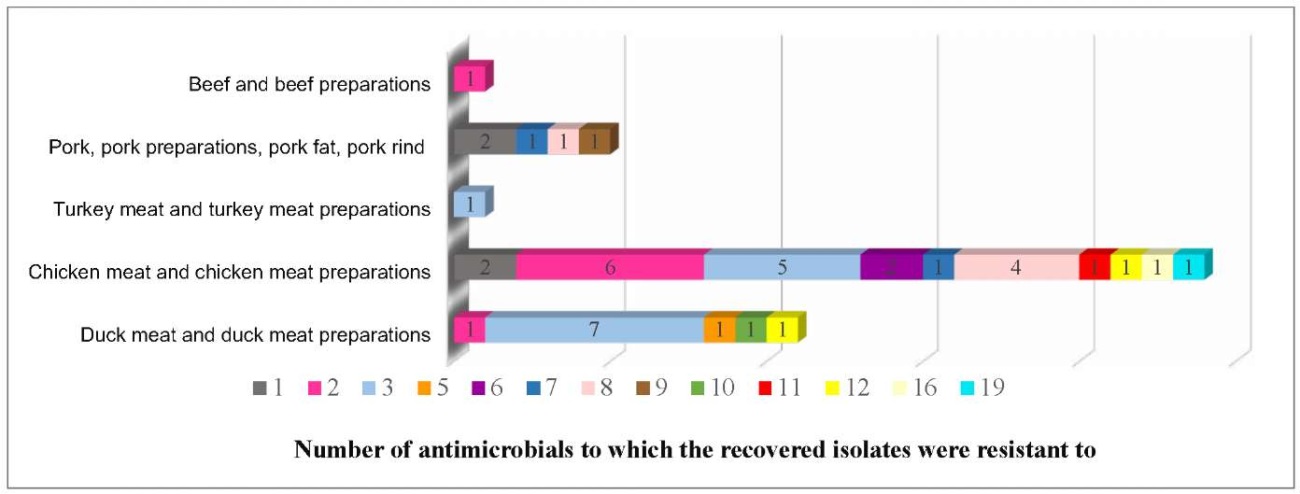
Fig. 5. Number of Salmonella spp. isolates demonstrating multiple antimicrobial resistance recovered from animal product samples in 2022–2024
The isolate resistant to 6 antibiotics and the isolate resistant to 19 out of tested 21 antibiotics were detected during the study. Both isolates belonged to O:4 (B) group and were detected in chicken meat.
According to the official report “On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2024”, the majority of resistant Salmonella spp. were detected in poultry meat, eggs and products thereof (97.2%) and in meat and meat products (95.1%) 5.
According to some authors, most of the Salmonella spp. isolated from the products are resistant to at least one class of antimicrobials. For example, the ECDC informs that in the European Union Salmonella-infected people often demonstrate resistance to antimicrobials, therewith more than 20% of such patients demonstrate resistance to at least three classes of antimicrobials. Moreover, according to the WHO, resistance to fluoroquinolones and cephalosporins is increasing every year [1][18][21][22].
Currently, reports on multiple resistance of Salmonella spp. are appearing more frequently. Tests of Salmonella spp. detected in pig products have shown their high resistance to tetracycline, streptomycin, and sulfamethoxazole/trimethoprim [23][24].
Tests of Salmonella spp. isolates recovered from pig products performed during this study showed similar results: 80% of the isolates demonstrated high resistance to erythromycin.
The Federal Service for Supervision of Consumer Rights Protection and Human Welfare in its official report for 2024 indicated that Salmonella spp. demonstrated resistance to one or more antimicrobials with the highest resistance observed to tetracycline, ciprofloxacin and sulfamethoxazole/trimethoprim.
Thus, excessive antibiotic use fuels multiple antimicrobial resistance leading to longer and more expensive treatment, as well as fatal outcomes and economic losses, thereby posing a great threat [25][26].
Figure 6 shows dynamics of increase in Salmonella spp. isolates resistant to antimicrobials of the same class in 2022 to 2024.
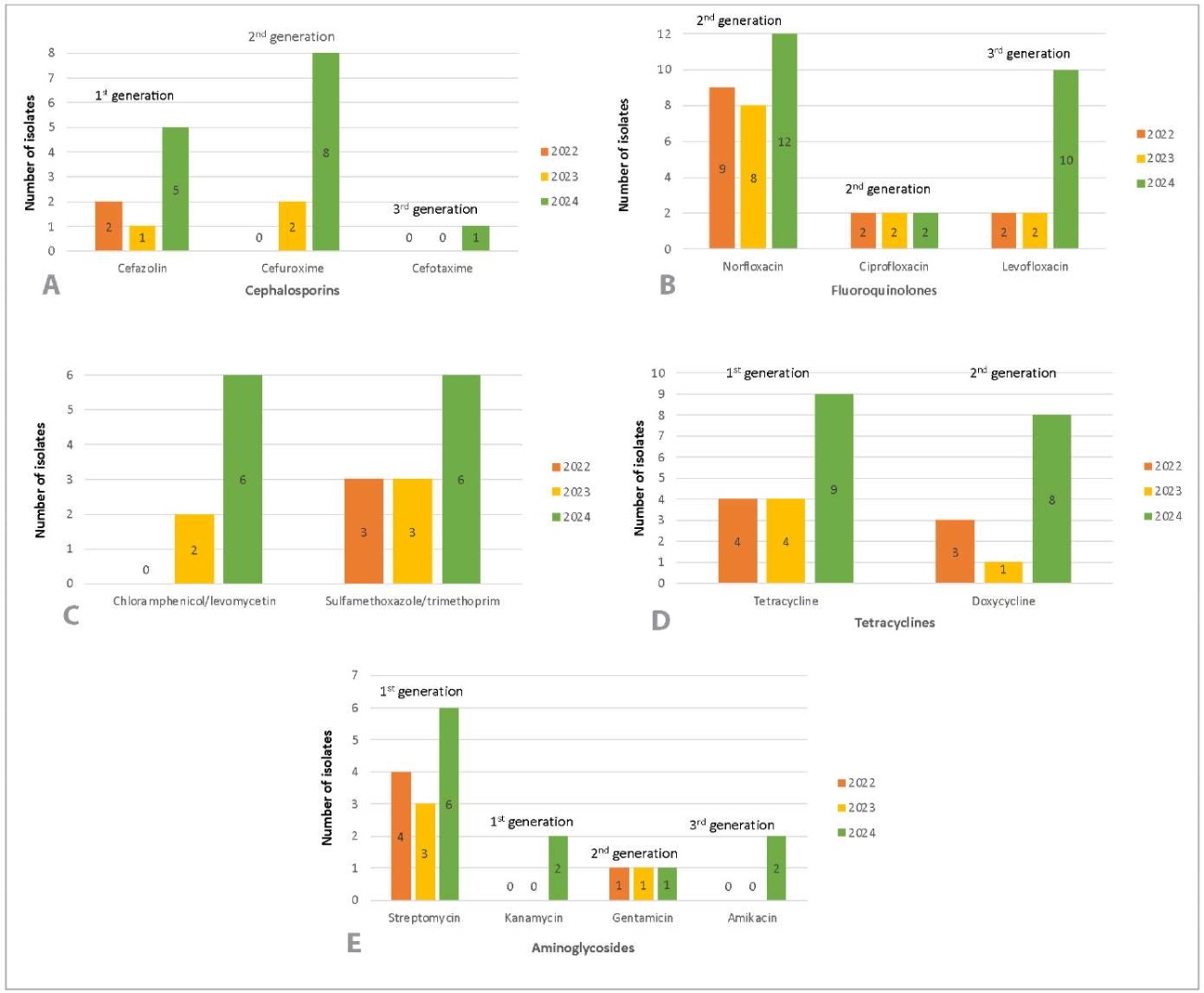
Fig. 6. Resistance of Salmonella spp. isolates to the following antibiotics: A – cephalosporins, B – fluoroquinolones, C – chloramphenicol/levomycetin and sulfamethoxazole/trimethoprim, D – tetracyclines, E – aminoglycosides
The use of cephalosporins, in particular the third generation cephalosporins, for salmonellosis treatment has long been the most promising option owing to their high efficacy against Salmonella spp., resistance to bacterial beta-lactamases, excellent bioavailability, and favourable safety profile, especially in short-term treatment regimens. Fluoroquinolones are also effective antimicrobials against salmonellosis owing to their good cellular penetration. However, Salmonella spp. are increasingly developing resistance to this class of antimicrobials worldwide [27][28].
The study results (Fig. 6A) have shown that resistance of Salmonella spp. to cephalosporins has increased since 2022: resistance to the first generation cephalosporin (cefazolin) has increased by 18.0% (5 isolates (31.3%) out of 16 recovered isolates were resistant in 2024), resistance to the second generation cephalosporin (cefuroxime) has increased by 50.0% (8 isolates (50.0%) out of 16 recovered isolates were resistant in 2024) and resistance to the third generation cephalosporin (cefotaxime) has increased by 6.3% (1 isolate (6.3%) out of 16 recovered isolates was resistant in 2024, previously no resistant isolates were detected).
Figure 6B shows the similar results for norfloxacin (second generation fluoroquinolone): the resistance has increased by 15.0% (12 isolates (75.0%) out of 16 recovered isolates were resistant in 2024), as well as resistance to levofloxacin (third generation fluoroquinolone) has increased by 49.2% (10 isolates (62.5%) out of 16 recovered isolates were resistant in 2024).
The resistance of isolates to chloramphenicol/levomycetin (increased by 37.5%) and sulfamethoxazole/trimethoprim (increased by 17.5%) has changed in 2022–2024 (Fig. 6C).
High increase in resistance to tetracyclines was detected in 2022–2024 (Fig. 6D): resistance to tetracycline (first generation) increased by 29.6% (9 isolates (56.3%) out of 16 recovered isolates were resistant in 2024), resistance to doxycycline (second generation) increased by 30.0% (8 isolates (50.0%) out of 16 recovered isolates were resistant in 2024).
At the same time, an increase in aminoglycosides resistance was observed (Fig. 6E). Thus, amikacin (third generation) and kanamycin (first generation), resistance has increased by 12.5% (2 isolates (12.5%) out of 16 recovered isolates were resistant in 2024). Streptomycin (first generation) resistance increased by 10.8% (6 isolates (37.5%) out of 16 recovered isolates were resistant in 2024).
No antimicrobial resistance genes, blaCTX-M, blaOXA10, blaDHA, blaGES, blaKPC, blaOXA48-like, blaNDM, or blaVIM, were found in the recovered Salmonella spp. isolates during the study.
CONCLUSION
Forty-two Salmonella spp. isolates were detected during the study; the predominant serovars were as follows: S. Enteritidis – 14 (33.3%), S. Blegdam – 3 (7.1%), S. Derby – 2 (4.8%).
Significant spread of resistance, including multiple resistance, has been shown. Salmonella spp. isolates demonstrated maximum resistance to erythromycin (80.9%), benzylpenicillin (78.6%), norfloxacin (69.0%) and tetracycline (40.5%). All Salmonella spp. isolates were susceptible to meropenem and imipenem.
Most isolates of Salmonella spp. demonstrated resistance to three antimicrobials at once (31.0%), and one isolate was also found to be resistant to 19 out of 21 antimicrobials used for the study.
In addition, an increase in resistance of Salmonella spp. isolates to the following antimicrobials was shown: cephalosporins – resistance to the first generation (cefazolin) increased by 18.0%, resistance to the second generation (cefuroxime) increased by 50.0%, resistance to the third generation (cefotaxime) increased by 6.3%; fluoroquinolones – resistance to the second generation (norfloxacin) increased by 15.0%, resistance to the third generation (levofloxacin) increased by 49.2%; tetracyclines – resistance to the first generation (tetracycline) increased by 29.6%, resistance to the second generation (doxycycline) increased by 30.0%.
Besides, resistance of isolates to chloramphenicol/levomycetin increased by 37.5% and to sulfamethoxazole/trimethoprim increased by 17.5% during the study period, 2022 –2024.
Resistance to aminoglycosides has also increased: resistance to amikacin (third generation) and kanamycin (first generation) has increased by 12.5%; resistance to streptomycin (first generation) has increased by 10.8%.
No antimicrobial resistance genes (blaCTX-M, blaOXA10, blaDHA, blaGES, blaKPC, blaOXA48-like, blaNDM, blaVIM) have been detected in the recovered Salmonella spp.
Continuous monitoring of animal product quality enables prompt detection of shifts in bacterial populations, facilitating the development of effective strategies for reducing the transmission of resistant strains and genes to humans. However, monitoring for antimicrobial resistance trends in isolated strains supports rational antibiotic use in veterinary and clinical medicine. This is essential for effective salmonellosis surveillance under the One Health framework.
Contribution of the authors: Akulich O. А. – testing, obtained data analysis and interpretation, paper text preparation, figure creation; Shadrova N. B. – formulation of key goals and objectives, paper text editing and approval of final paper text; Denisova G. S. – conceptualization, formulation of key goals and objectives, paper text editing.
Вклад авторов: Акулич О. А. – проведение исследований, анализ и интерпретация полученных данных, подготовка текста статьи, создание рисунков; Шадрова Н. Б. – формулировка ключевых целей и задач, редактирование текста статьи и утверждение окончательного варианта; Денисова Г. С. – формирование идеи, формулировка ключевых целей и задач, редактирование текста статьи.
1. https://www.rospotrebnadzor.ru/documents/details.php?ELEMENT_ID=21796 (in Russ.)
2. https://www.rospotrebnadzor.ru/upload/iblock/b50/t4kqksh4b12a2iwjnha29922vu7naki5/GD-SEB.pdf (in Russ.)
3. https://www.rospotrebnadzor.ru/documents/details.php?ELEMENT_ID=27779 (in Russ.)
4. https://www.rospotrebnadzor.ru/upload/iblock/b8a/u6lsxjabw032jkdf837nlaezxu3ue09m/GD_SEB.pdf (in Russ.)
5. https://www.rospotrebnadzor.ru/upload/iblock/b8a/u6lsxjabw032jkdf837nlaezxu3ue09m/GD_SEB.pdf (in Russ.)
References
1. WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization; 2024. 56 р. https://iris.who.int/bitstream/handle/10665/376776/9789240093461-eng.pdf?sequence=1
2. World Health Organization.Ten threats to global health in 2019. https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019
3. World health statistics 2024: monitoring health for the SDGs, Sustainable Development Goals. Geneva: World Health Organization; 2024. 86 р. https://iris.who.int/bitstream/handle/10665/376869/9789240094703-eng.pdf
4. Tarín-Pelló A., Suay-García B., Pérez-Gracia M.-T. Antibiotic resistant bacteria: current situation and treatment options to accelerate the development of a new antimicrobial arsenal. Expert Review of Anti-Infective Therapy. 2022; 20 (8): 1095–1108. https://doi.org/10.1080/14787210.2022.2078308
5. Di K. N., Pham D. T., Tee T. S., Binh Q. A., Nguyen T. C. Antibiotic usage and resistance in animal production in Vietnam: a review of existing literature. Tropical Animal Health and Production. 2021; 53 (3):340. https://doi.org/10.1007/s11250-021-02780-6
6. Mukhina E., Artemieva M., Sakunts L., Tozhiboeva B. The social problem of antibiotic resistance. Universum: Medicine & Pharmacology. 2017; (6). https://7universum.com/ru/med/archive/item/4898 (in Russ.)
7. Manyi-Loh C., Mamphweli S., Meyer E., Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules. 2018; 23 (4):795. https://doi.org/10.3390/molecules23040795
8. Aleksandrowicz A., Carolak E., Dutkiewicz A., Błachut A., Waszczuk W., Grzymajlo K. Better together – Salmonella biofilm-associated antibiotic resistance. Gut Microbes. 2023; 15 (1):2229937. https://doi.org/10.1080/19490976.2023.2229937
9. Wang B. X., Butler D. S. С., Hamblin M., Monack D. M. One species, different diseases: the unique molecular mechanisms that underlie the pathogenesis of typhoidal Salmonella infections. Current Opinion in Microbiology. 2023; 72:102262. https://doi.org/10.1016/j.mib.2022.102262
10. Li S., He Y., Mann D. A., Deng X. Global spread of Salmonella Enteritidis via centralized sourcing and international trade of poultry breeding stocks. Nature Communications. 2021; 12:5109. https://doi.org/10.1038/s41467-021-25319-7
11. Egorova S. A., Kaftyreva L. A., Pomazanov V. V. Current trends in the development of resistance to clinically significant antibiotics in Salmonella (review of literature). Russian Clinical LaboratoryDiagnostics. 2020; 65 (5): 308–315. http://dx.doi.org/10.18821/0869-2084-2020-65-5-308-315 (in Russ.)
12. World Animal Protection: Global public health cost of antimicrobial resistance related to antibiotic use on factory farms. https://www.world-animalprotection.org.in/globalassets/pdfs/reports/english/global-public-health-technical-report.pdf
13. WHO estimates of the global burden of foodborne diseases: food-borne disease burden epidemiology reference group 2007–2015. Geneva: World Health Organization; 2015. 254 p. https://iris.who.int/bitstream/handle/10665/199350/9789241565165_eng.pdf?sequence=1
14. Wu S., Hulme J. P. Recent advances in the detection of antibiotic and multi-drug resistant Salmonella: an update. International Journal of Molecular Sciences. 2021; 22 (7):3499. https://doi.org/10.3390/ijms22073499
15. Determination of the sensitivity of microorganisms to antimicrobial drugs: Russian recommendations. Version 2025-01. Smolensk: Smolensk State Medical University; Interregional Association for Clinical Microbiology and Antimicrobial Chemotherapy; 2025. 208 р. https://www.antibiotic.ru/library/ocmap2025 (in Russ.)
16. European Committee on Antimicrobial Susceptibility Testing. Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST. Version 15.0, valid from 2025-01-01. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/QC/v_15.0_EUCAST_QC_tables_routine_and_extended_QC.pdf
17. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 35th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute; 2025. 428 p.
18. Salmonellosis. In: European Centre for Disease Prevention and Control. Annual Epidemiological Report for 2022. Stockholm: ECDC; 2024. https://www.ecdc.europa.eu/en/publications-data/salmonellosis-annual-epidemiological-report-2022
19. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2022–2023. EFSA Journal. 2025; 23 (3):e9237. https://doi.org/10.2903/j.efsa.2025.9237
20. Vitkova O. N., Belousov V. I., Ivanova O. E., Bazarbaev S. B. Study of antibiotic resistance of Salmonella isolates from animals and fodders of animal origin on the territory of the Russian Federation. Veterinaria Kubani. 2015; (2): 11–15. https://elibrary.ru/tppjdx (in Russ.)
21. Rakitin A. L., Yushina Y. K, Zaiko E. V., Bataeva D. S., Kuznetsova O. A., Semenova A. A., et al. Evaluation of antibiotic resistance of Salmonella serotypes and whole-genome sequencing of multiresistant strains isolated from food products in Russia. Antibiotics. 2022; 11 (1):1. https://doi.org/10.3390/antibiotics11010001
22. Mendybaeva A. M., Ruzauskas M., Aleshina Yu. E., Alieva G. K., Mukanov G. B., Ryshchanova R. M. Opportunistic and pathogenic microflora antibiotics resistance risk assessment extracted from animal products. Bulletin KrasSAU. 2022; (2): 147–156. https://doi.org/10.36718/1819-4036-2022-2-147-156 (in Russ.)
23. Patchanee P., Tansiricharoenkul K., Buawiratlert T., Wiratsudakul A., Angchokchatchawal K., Yamsakul P., et al. Salmonella in pork retail outlets and dissemination of its pulsotypes through pig production chain in Chiang Mai and surrounding areas, Thailand. Preventive Veterinary Medicine. 2016; 130: 99–105. https://doi.org/10.1016/j.prevetmed.2016.06.013
24. Possebon F. S., Tiba Casas M. R., Nero L. A., Yamatogi R. S., Araújo J. P. Jr., Pinto J. P. A. N. Prevalence, antibiotic resistance, PFGE and MLST characterization of Salmonella in swine mesenteric lymph nodes. Preventive Veterinary Medicine. 2020; 179:105024. https://doi.org/10.1016/j.prevetmed.2020.105024
25. Solov’yeva A. S., Shubin F. N., Kuznetsova N. A. Antibiotic resistance of Salmonella enteritidis sticks allocated in the Far Eastern and Siberian Federal Districts. Health. Medical ecology. Science. 2017; (5): 15–21. https://doi.org/10.5281/zenodo.1115444 (in Russ.)
26. Baquero F. Threats of antibiotic resistance: an obliged reappraisal. International Microbiology. 2021; 24 (4): 499–506. https://doi.org/10.1007/s10123-021-00184-y
27. Wen S. C. H., Best E., Nourse C. Non-typhoidal Salmonella infections in children: review of literature and recommendations for management. Journal of Paediatrics and Child Health. 2017; 53 (10): 936–941. https://doi.org/10.1111/jpc.13585
28. Konyali D., Guzel M., SoyerY. Genomic characterization of Salmonella enterica resistant to cephalosporin, quinolones, and macrolides. Current Microbiology. 2023; 80 (11):344. https://doi.org/10.1007/s00284-023-03458-y
About the Authors
O. A. AkulichRussian Federation
Olga А. Akulich - Postgraduate Student, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
N. B. Shadrova
Russian Federation
Natalya B. Shadrova - Cand. Sci. (Biology), Head of Department for Microbiological Testing, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
G. S. Denisova
Russian Federation
Galina S. Denisova - Cand. Sci. (Biology), Head of the Vladimir Testing Centre, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
Review
For citations:
Akulich O.A., Shadrova N.B., Denisova G.S. Antimicrobial resistance of Salmonella spp. detected in animal products in 2022–2024. Veterinary Science Today. 2025;14(3):310-318. https://doi.org/10.29326/2304-196X-2025-14-3-310-318



































