Scroll to:
Phylogenetic analysis of rabies virus isolates recovered from animals in Volgograd Oblast
https://doi.org/10.29326/2304-196X-2025-14-3-241-248
Abstract
Introduction. The Lower Volga region, including the Volgograd Oblast, remains one of Russia’s most rabies-affected areas to date. Data on the genetic diversity of rabies viruses (RABVs) currently circulating in the Volgograd Oblast are insufficient, making phylogenetic analysis of RABV isolates from this region a relevant scientific objective.
Objective. The study aims to conduct a phylogenetic analysis of current RABV isolates recovered from animals in the Volgograd Oblast, based on the full-length nucleoprotein gene sequence.
Materials and methods. Brain tissue samples from animals diagnosed with rabies were used. The obtained nucleotide sequences of the RABV nucleoprotein gene were analyzed using the Bayesian strict molecular clock method. The spatial distribution of RABV isolates was described using Natural Earth physical map.
Results. Full-length nucleoprotein gene sequencing was performed for 13 RABV isolates collected in the Volgograd Oblast. Phylogenetic analysis revealed that the RABV population in this region comprises distinct genetic variants of genetic group C, formed at different times. Genetic relationship to the isolates from Kazakhstan, Ukraine, Moldova and Central/Southern Russia indicates intensive RABV circulation in Southern European Russia. Notably, distinct virus variants were detected on the left and right banks of the Volga River.
Conclusion. All studied RABV isolates collected in the Volgograd Oblast belonged to genetic group C and exhibited high genetic diversity among variants.
For citations:
Chupin S.A., Chernyshova E.V., Chernyshev R.S., Gruzdev K.N., Spiridonov A.N., Varkentin A.V., Nazarov N.A., Zarva I.D., Botvinkin A.D. Phylogenetic analysis of rabies virus isolates recovered from animals in Volgograd Oblast. Veterinary Science Today. 2025;14(3):241-248. https://doi.org/10.29326/2304-196X-2025-14-3-241-248
INTRODUCTION
The application of molecular genetic studies in epizootology has significantly enhanced surveillance capabilities for zoonotic infections. Data on intra-species diversity and genetic lineage relationships of pathogens, obtained through phylogenetic analysis of both individual gene sequences and complete genomes, provide critical insights into pathogen origins, spatial distribution patterns, and their significance in human and veterinary pathology. These advances have established and continue to develop two emerging fields: molecular epidemiology and genomic epidemiological surveillance [1][2].
Multiple genetically distinct rabies virus (RABV) groups with distinct geographical distributions have been identified in the Russian Federation [3][4][5][6][7]. Representatives of genetic group C ("steppe") [3] demonstrate the widest distribution, currently spanning territories from Eastern Europe through southern Siberia to Kazakhstan and northeastern China [6][8][9][10][11]. Since the mid-20th century, the red fox (Vulpes vulpes) has served as the primary RABV reservoir in this region. Current epidemiological models suggest that fox-mediated rabies epizootics originated in 1939 in East Prussia (Polish Corridor), subsequently spreading westward to France and Belgium and eastward to the Baltics, Ukraine, Belarus and western Russia within 20–30 years [12][13]. Cartographic analyses clearly document this progressive expansion of fox rabies across the European territories of the former USSR [14]. By the early 21st century, these epizootics had reached previously rabies-free Siberian regions, including Krasnoyarsk Krai and Transbaikalia, after decades of absence [7][15].
A wild rabies outbreak was documented in the Volga River Delta in 1942 [13]. This region, along with East Prussia, is believed to have been one of the epicenters of fox rabies spread in the USSR [13][16]. To this day, the Lower Volga region, including the Volgograd Oblast, remains one of Russia’s most rabies-endemic areas [7][16]. Nearly the entire territory of the Volgograd Oblast is enzootic for the disease [17][18]. Although the fox is considered the RABV primary reservoir in this region, up to 38% of all recorded cases in recent decades have involved dogs and cats, while wild animals accounted for no more than 25% [19]. Notably, in 4 out of 6 human rabies cases reported after 2000, transmission occurred via dogs and cats [7][20]. To date, nucleotide sequences of the nucleoprotein gene (N gene) have been published for only two RABV isolates from animals in the Volgograd Oblast, both belonging to genetic group C [3]. Given the limited data on the genetic diversity of current RABVs in the region, a phylogenetic analysis of the virus isolates from the Volgograd Oblast remains a relevant and necessary research objective.
The aim of the study was to conduct a phylogenetic analysis of currently circulating RABV isolates recovered from animals in the Volgograd Oblast, based on the complete nucleoprotein (N) gene sequence.
MATERIALS AND METHODS
A total of 13 RABV isolates collected from animals in the Volgograd Oblast in 2018–2021 were studied (Table 1, Fig. 1).
Table 1
Information on RABV isolates recovered from animals in the Volgograd Oblast
|
№ |
Isolate full name |
Coordinates of populated localitites |
Year |
Host animal |
GenBank accession number |
|
|
latitude |
longitude |
|||||
|
1 |
490_2/2018/Volgograd |
50.19760 |
42.69182 |
2018 |
dog |
OP311892 |
|
2 |
490_4/2018/Volgograd |
49.98861 |
46.68835 |
2018 |
dog |
OP311893 |
|
3 |
1563/2018/Volgograd |
50.99677 |
44.35670 |
2018 |
dog |
OP311840 |
|
4 |
1299/138/2021/Volgograd |
51.07931 |
42.51306 |
2021 |
cattle |
OP311869 |
|
5 |
1299/139/2021/Volgograd |
50.08529 |
45.40216 |
2021 |
dog |
OP311870 |
|
6 |
1299/141/2021/Volgograd |
48.38944 |
42.35912 |
2021 |
dog |
OP311871 |
|
7 |
1299/142/2021/Volgograd |
48.09285 |
42.59016 |
2021 |
cattle |
OP311872 |
|
8 |
1299/143/2021/Volgograd |
50.24182 |
41.83726 |
2021 |
dog |
OP311873 |
|
9 |
1299/144/2021/Volgograd |
49.85802 |
42.33575 |
2021 |
dog |
OP311874 |
|
10 |
1299/145/2021/Volgograd |
49.35021 |
45.07359 |
2021 |
marten* |
OP311875 |
|
11 |
1299/146/2021/Volgograd |
49.73250 |
43.48296 |
2021 |
cattle |
OP311876 |
|
12 |
1299/147/2021/Volgograd |
48.24536 |
42.64595 |
2021 |
cat |
OP311877 |
|
13 |
1299/148/2021/Volgograd |
49.85802 |
42.33575 |
2021 |
wolf** |
OP311878 |
|
* marten – Martes sp.; ** wolf – Canis lupus. |
||||||
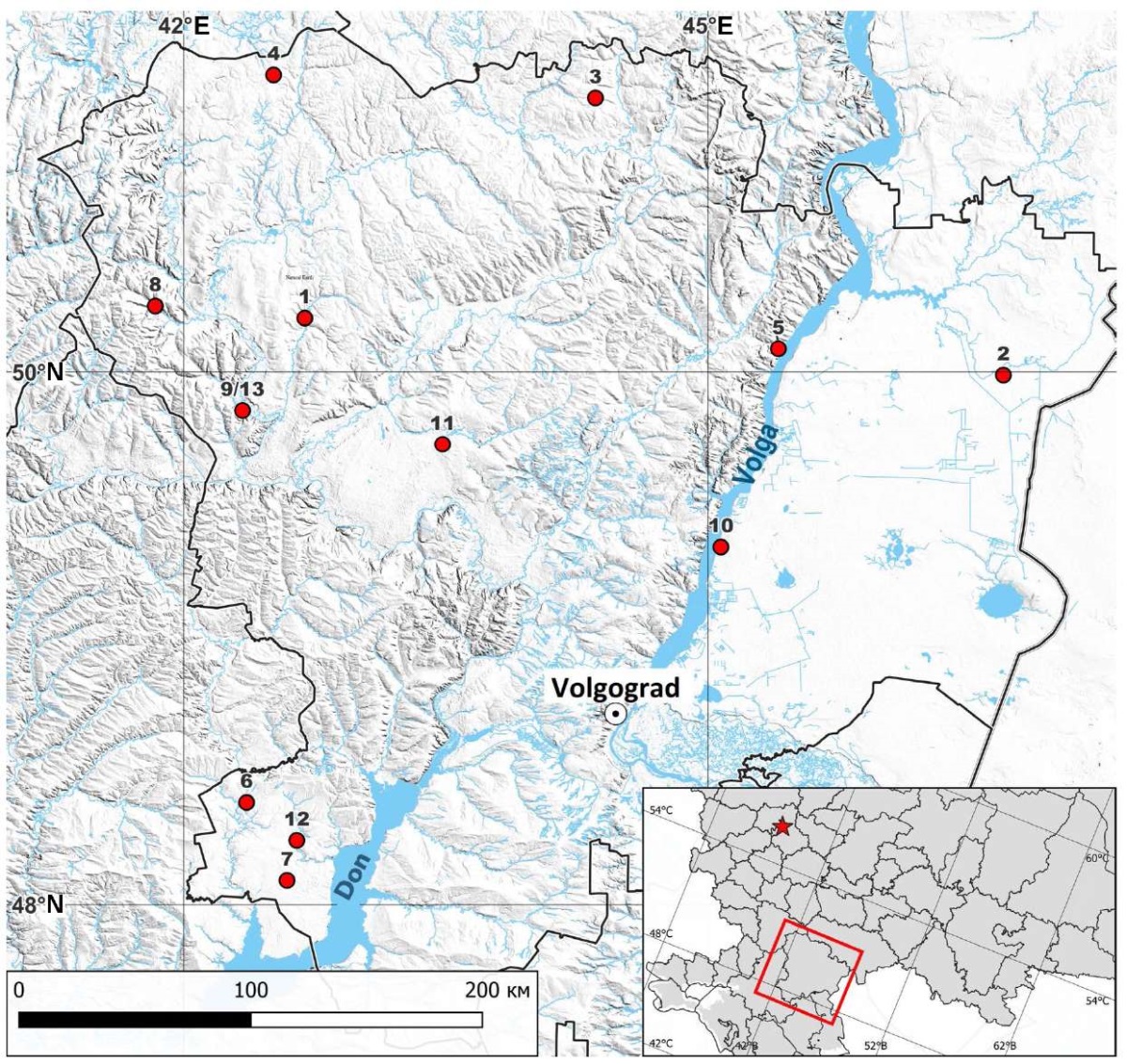
Fig. 1. Distribution of RABV isolates collected from animals in the Volgograd Oblast (the isolates listed in Table 1 are numbered)
RNA was extracted from animal brain tissue, and two overlapping genome fragments containing the full-length nucleoprotein (N) gene of RABV were amplified by reverse transcription polymerase chain reaction. Following purification, the fragments were subjected to Sanger sequencing as previously described [5]. The resulting sequences were deposited in the international GenBank database (accession numbers provided in Table 1).
The phylogenetic analysis was performed using the Bayesian approach in the Bayesian Evolutionary Analysis Sampling Trees (BEAST X) software package [21]. A model suitable for constructing a phylogenetic tree was preliminarily evaluated using the MEGA X program, with the Bayesian information criterion (BIC) and the corrected Akaike information criterion (AICc) determined. Based on the test results, the Hasegawa-Kishino-Yano model with a 4-category gamma distribution (HKY + G; BIC = 21,360.059; AICc = 17,664.682) was selected [22]. A phylogenetic tree reflecting the posterior probability of the formation time of internal (ancestral) and terminal nodes with a 95% confidence interval (95% CI) was constructed using the strict molecular clock method, with heterochronic calibration to the year of sample isolation, a random initial tree model with a constant population size, and a Markov chain Monte Carlo (MCMC) chain length of 1 × 10⁸ with sampling every 1 × 10⁵ steps. The posterior prognostic verification was performed for all taxa belonging to the genetic group C of the RABV.
The reliability of the MCMC was assessed using the Tracer program by analyzing the BEAST X output data [23].
The tree was annotated using the TreeAnnotator utility and the settings of outlier removal (burn-in = 10%, 10,000,000 states), maximum clade confidence (MCC), and maintaining a given length.
Visualization of the phylogenetic tree was performed using the FigTree v.1.4.5 program [24].
The QGIS 3.2.1 program and the Natural Earth electronic landscape-geographic map were used for cartography. The points were plotted on the map according to the geographic coordinates of settlements where rabies cases were detected.
RESULTS AND DISCUSSION
The full-length nucleotide sequence of the N gene (1353 nt) was determined for 13 RABV field isolates collected from animals in ten raions of the Volgograd Oblast in 2018–2021. The map shows the locations where isolates were detected (Fig. 1).
According to preliminary analysis, all studied isolates belong to RABV genetic group C. Therefore, to construct the phylogenetic tree, full-length sequences of all available isolates from this group identified in the Russian Federation with known collection years were retrieved from the GenBank database. Additionally, BLAST (Basic Local Alignment Search Tool, https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis of the studied sequences revealed closely related isolates from Kazakhstan, Moldova, Poland, and Ukraine, which were also included in the phylogenetic analysis. To clearly represent group C, several isolates from other genetic groups circulating in Europe and European Russia were included as genetic variants (excluding Arctic-related clade representatives due to their distant genetic relationship). In total, 178 full-length N gene sequences were analyzed, including 121 sequences from genetic group C. For improved figure readability, the isolate groups phylogenetically distant from the tested ones were collapsed. The resulting phylogenetic tree is presented in Figure 2.
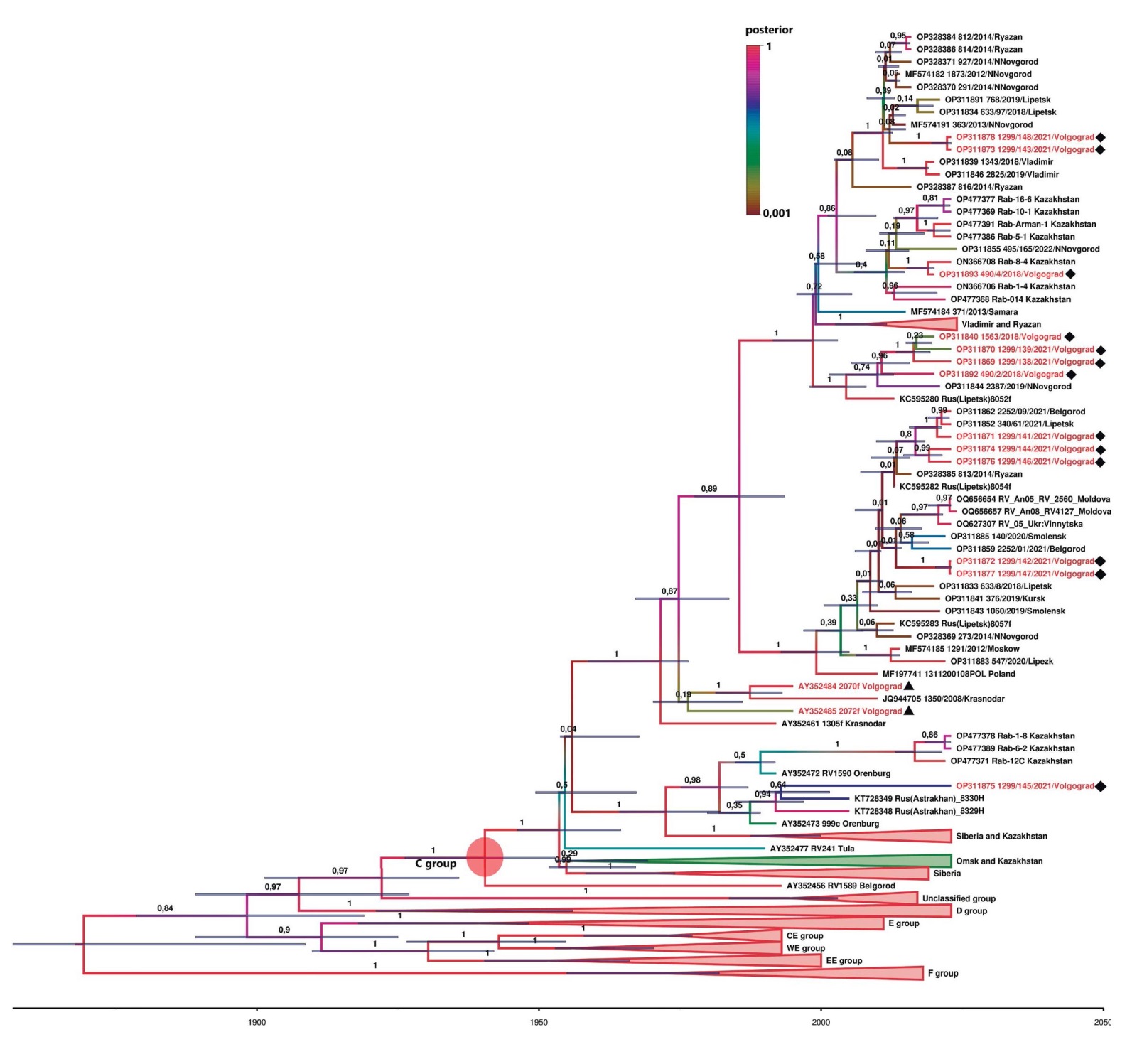
Fig. 2. Phylogenetic tree constructed using the full-length nucleotide sequence of RABV N gene and a strict molecular clock algorithm. Isolates collected from animals in the Volgograd Oblast are highlighted in red. Numbers (0–1) indicate posterior probability values for specific groups; the same parameter is indicated with the color of branches (see color scale for interpretation). The horizontal timeline at the bottom shows the estimated years of the most recent common ancestor for this or that group, with blue bars at nodes representing 95% confidence intervals for this value. The pink circle marks the most recent common ancestor of genetic group C. Black diamonds denote the studied isolates, while triangles represent two previously characterized isolates from the Volgograd Oblast
According to the tree topology, all the studied isolates belong to the genetic group C, the modern range of which within the Russian Federation extends from the western borders and regions of the central zone of the European part of the country to the steppe and forest-steppe territories of Siberia, everywhere going beyond the borders of Russia in the south [3][6][5][11]. RABV of genetic lineage C was isolated from both wild carnivores and domestic and farm animals in the Volgograd Oblast.
All studied isolates are unique compared to previously identified isolates, including those from the Volgograd Oblast, and form several groups with a high posterior probability estimate (PPE) with each other and with previously studied isolates from Russian and foreign regions.
Thus, isolate 490_4/2018/Volgograd shows significant similarity (99.85%) with isolate Rab-8-4, identified in the Turkestan Oblast of Kazakhstan. The estimated time of evidence of the most recent common ancestor (MRCA) for this group is 2015.9 (95% CI – from 2012.1 to 2016.93), the PPE of the group is 1.0.
Isolates 1299/143/2021/Volgograd and 1299/148/2021/Volgograd show the greatest relationship with isolates previously identified in the Vladimir, Lipetsk, Nizhny Novgorod and Ryazan Oblasts (more than 98.67%). The MRCA for this group is 2007.95 (95% CI – from 2002.48 to 2009.67), the PPE of the group is 1.0.
The group of isolates (490_2/2018/Volgograd, 1299/138/2021/Volgograd, 1299/139/2021/Volgograd, 1563/2018/Volgograd) shows the greatest affinity (more than 98.9%) with the isolates previously identified in the Nizhny Novgorod and Lipetsk Oblasts (but with others than in previous cases). The MRCA for this group is 2001.43 (95% CI – from 1994.02 to 2002.65), the PPE of the group is 1.0.
Another five isolates from the Volgograd Oblast (1299/142/2021/Volgograd, 1299/147/2021/Volgograd, 1299/141/2021/Volgograd, 1299/144/2021/Volgograd, 1299/146/2021/Volgograd) form a group with a high PPE (1.0) with isolates from the Belgorod, Lipetsk, Ryazan, Smolensk, Kursk, Nizhny Novgorod, Moscow Oblasts, as well as with isolates identified in other countries – Moldova, Ukraine and Poland. It is noteworthy that this relatively genetically dense group occupies an extremely vast area, stretching at least 1,000 km from north to south and 2,000 km from west to east. The MRCA for this group was 1996.13 (95% CI from 1989.12 to 2003.42).
Isolate 1299/145/2021/Volgograd genetically differs significantly from other Volgograd isolates and shows the greatest (although not very close) relationship with isolates identified in the Astrakhan Oblast (98.74%). It forms a group supported by a high PPE (0.98) with RABV isolates identified in the Orenburg and Astrakhan Oblasts, as well as in Kazakhstan. The MRCA for this group is 1978.98 (1974.35–1983.97).
Two isolates (2070f and 2072f) identified in the Volgograd Oblast by I. V. Kuzmin et al. [3] in 2003 differ significantly from the studied isolates from this region. Isolate 2070f forms a group with isolate 1350/2008/Krasnodar from the Krasnodar Krai (PPE – 1.0; MRCA – 1984.33; 95% CI –1977.17–1989.78), and isolate 2072f, although having a genetic relationship with isolate 2070f (99.1%), but their group has a very low posterior probability support – 0.19.
The number of analyzed isolates collected from various animal groups generally reflects the morbidity structure observed during 2020–2024. Over 50% of these isolates were derived from canine and feline specimens (Table 2).
Table 2
Data on rabies confirmation in animals from the Volgograd Region (2020–2024*) and sources of virus isolates for phylogenetic analysis
|
Animal categories |
Confirmed rabies cases |
Number of tested isolates |
||
|
n |
% |
n |
% |
|
|
Livestock (cattle, small ruminants, horses) |
42 |
23.3 |
3 |
23.1 |
|
Domestic carnivores (dogs, cats) |
109 |
60.6 |
8 |
61.5 |
|
Wild carnivores (foxes, wolves, etc.) |
29 |
16.1 |
2 |
15.4 |
|
Total |
180 |
100 |
13 |
100 |
|
* Data for 2024 (9 months). |
||||
Thus, the key characteristic of the RABV population in the Volgograd Oblast is its remarkable heterogeneity, possibly the highest among well-studied regions to date. At the same time, nearly all representatives of the Volgograd RABV population have close genetic relatives in other regions of Russia and abroad, suggesting active viral spread. According to published data, increased epizootic activity in the Volgograd Oblast was recorded in 1988, 1991, 1997, 1998, and 2001 [17]. The Volgograd RABV population comprises multiple genetic variants, which emerged at different times, as evidenced by the estimated age of their last common ancestors. It is plausible that peaks in epizootic activity coincide with the introduction of new viral variants. Following their spread, these variants establish themselves among susceptible hosts in the region, continue to evolve, and ultimately contribute to the highly diverse genetic profile observed in the current population.
The localization features of different RABV subgroups are noteworthy. The Volgograd Oblast occupies the central part of the southeast of the Russian (East European) Plain within three natural zones: forest-steppe, steppe and semi-desert [25]. Isolates collected in the forest-steppe and steppe regions of the northern part of the Volgograd Oblast on the right bank of the Volga (490_2/2018/Volgograd, 1563/2018/Volgograd, 1299/138/2021/Volgograd, 1299/139/2021/Volgograd, 1299/143/2021/Volgograd, 1299/144/2021/Volgograd, 1299/148/2021/Volgograd) are genetically related to isolates from the Middle Volga and central regions of Russia. This likely results from the absence of natural barriers to wild carnivore migration in northern directions, combined with high fox population densities in adjacent areas [26]. RABV isolates 1299/141/2021/Volgograd, 1299/142/2021/Volgograd, 1299/147/2021/Volgograd, which are similar to the isolates found in Ukraine, Moldova, Poland, and western/central regions of European Russia, were identified in the southwestern Volgograd Oblast on the right bank of the Tsimlyansk Reservoir. Isolates from the left bank of the Volga (490_4/2018/Volgograd, 1299/145/2021/Volgograd) show genetic similarity to strains from southern and eastern neighboring regions, reflecting shared landscape characteristics with the semi-deserts and dry steppes of Kazakhstan and Astrakhan Oblast. Major water barriers (Tsimlyansk and Volgograd Reservoirs and the Volga-Don Canal), which cross the entire region northeast to southwest, freeze for only 2–3 months annually [25] and consequently serve as effective natural barriers to rabies spread for most of the year. We obtained only two isolates from the left bank of the Volga, both distinct from the right-bank strains. Thus, isolate 1299/145/2021/Volgograd (recovered in the Bykovsky Raion), clusters with strains from Orenburg and Astrakhan Oblasts (exact origins unknown) and Kazakhstan. Isolate 490/4/2018/Volgograd (identified in the Pallasovsky Raion) shows closest relations to some Kazakhstan strains and one strain (being the only exception) from the Nizhny Novgorod Oblast. Notably, raions with highest animal rabies incidence (Nikolaevsky, Bykovsky, Leninsky, Oktyabrsky) [19] are all located on the left Volga bank. The Volga-Akhtuba floodplain appears to function as a significant ecological corridor connecting Volgograd and Astrakhan Oblasts, which is supported by the genetic similarity of the isolates from these areas (1299/145/2021/Volgograd). The limited number of studied isolates prevents definitive conclusions. However, these findings warrant attention for ongoing research and strategic planning of fox oral vaccination programs.
CONCLUSION
All studied RABV isolates collected from animals in the Volgograd Oblast belong to genetic group C. Within this lineage, the RABV population in this region demonstrates exceptionally high genetic diversity. The genetic relatedness of these isolates to strains from Kazakhstan, Ukraine, Moldova and central/southern Russian regions suggests intensive host-mediated viral movement. This epidemiological pattern requires careful consideration when planning and implementing anti-epidemic measures.
Contribution of the authors: Chupin S. A. – conceptualization; formulation and development of key objectives and tasks, conducting research; analysis and interpretation of obtained data; text preparation and editing; Chernyshova E. V. – responsibility for all aspects of the work, ensuring integrity of all parts of the article and its final version; Chernyshev R. S. – application of phylogenetic statistical methods, preparation of figures, discussion of results; Gruzdev K. N. – scientific consulting, editing of the manuscript; Spiridonov A. N. – collection of statistical data; Varkentin A. V. – collection of statistical data; Nazarov N. A. – scientific consulting; Zarva I. D. – figure preparation, mapping; Botvinkin A. D. – conceptualization; formulation and development of key objectives and tasks; analysis and interpretation of obtained data; text preparation and editing.
Вклад авторов: Чупин С. А. – формирование идеи, формулировка и развитие ключевых целей и задач, проведение исследований, анализ и интерпретация полученных данных, подготовка и редактирование текста; Чернышова Е. В. – принятие ответственности за все аспекты работы, целостность всех частей статьи и за ее окончательный вариант; Чернышев Р. С. – применение статистических методов филогенетического анализа, подготовка рисунков, обсуждение результатов; Груздев К. Н. – научное консультирование, редактирование текста статьи; Спиридонов А. Н. – сбор статистических данных; Варкентин А. В. – сбор статистических данных; Назаров Н. А. – научное консультирование; Зарва И. Д. – подготовка рисунков, картография; Ботвинкин А. Д. – формирование идеи, формулировка и развитие ключевых целей и задач, анализ и интерпретация полученных данных, подготовка и редактирование текста.
References
1. Zhebrun A. B. From molecular to genomic and metagenomic epidemiology. Journal of Microbiology, Epidemiology and Immunobiology. 2014; 91 (3): 91–100. https://elibrary.ru/vobdlt (in Russ.)
2. Akimkin V. G., Semenenko T. A., Khafizov K. F., Ugleva S. V., Dubodelov D. V., Kolosovskaya E. N. Genomic surveillance strategy. Problems and perspectives. Journal of Microbiology, Epidemiology and Immunobiology. 2024; 101 (2): 163–172. https://doi.org/10.36233/0372-9311-507
3. Kuzmin I. V., Botvinkin A. D., McElhinney L. M., Smith J. S., Orciari L. A., Hughes G. J., et al. Molecular epidemiology of terrestrial rabies in the former Soviet Union. Journal of Wildlife Diseases. 2004; 40 (4): 617–631. https://doi.org/10.7589/0090-3558-40.4.617
4. Chupin S. A., Chernyshova E. V., Metlin A. E. Genetic characterization of the rabies virus field isolates detected in Russian Federation within the period 2008–2011. Problems of Virology. 2013; 58 (4): 44–49. https://elibrary.ru/rgqyth (in Russ.)
5. Chupin S. A., Sprygin A. V., Zinyakov N. G., Guseva N. A., Shcherbinin S. V., Korennoy F. I., et al. Phylogenetic characterization of rabies virus field isolates collected from animals in European Russian regions in 2009–2022. Microorganisms. 2023; 11 (10):2526. https://doi.org/10.3390/microorganisms11102526
6. Deviatkin A. A., Lukashev A. N., Poleshchuk E. M., Dedkov V. G., Tkachev S. E., Sidorov G. N., et al. The phylodynamics of the rabies virus in the Russian Federation. PLoS ONE. 2017; 12 (2):e0171855. https://doi.org/10.1371/journal.pone.0171855
7. Poleshchuk E. M., Sidorov G. N., Nashatyreva D. N., Gradoboeva E. A., Pakskina N. D., Popova I. V. Rabies in the Russian Federation: research and information newsletter. Omsk: Publishing Center KAN; 2019. 110 p. https://elibrary.ru/bouwsw (in Russ.)
8. Smreczak M., Orłowska A., Trębas P., Stolarek A., Freuling C., Müller T. Re-emergence of rabies in Mazowieckie Voivodeship, Poland, 2021. Zoonoses and Public Health. 2023; 70 (1): 111–116. https://doi.org/10.1111/zph.13005
9. Picard-Meyer E., Robardet E., Moroz D., Trotsenko Z., Drozhzhe Z., Biarnais M., et al. Molecular epidemiology of rabies in Ukraine. Archives of Virology. 2012; 157: 1689–1698. https://doi.org/10.1007/s00705-012-1351-6
10. Liu Y., Zhang S., Zhao J., Zhang F., Li N., Lian H., et al. Fox- and raccoon-dog-associated rabies outbreaks in northern China. Virologica Sinica. 2014; 29 (5): 308–310. https://doi.org/10.1007/s12250-014-3484-0
11. Yessembekova G. N., Xiao Sh., Abenov A., Karibaev T., Shevtsov A., Asylulan А., et al. Molecular epidemiological study of animal rabies in Kazakhstan. Journal of Integrative Agriculture. 2023; 22 (4): 1266–1275. https://doi.org/10.1016/j.jia.2022.11.011
12. Steck F., Wandeler A. The epidemiology of fox rabies in Europe. Epidemiologic Reviews. 1980; 2 (1): 71–96. https://doi.org/10.1093/oxfordjournals.epirev.a036227
13. Selimov M. A. Rabies. Moscow: Meditsina; 1978. 336 p. (in Russ.)
14. Botvinkin A., Kosenko M. Rabies in the European parts of Russia, Belarus and Ukraine. In: Historical Perspective of Rabies in Europe and the Mediterranean Basin. Eds. A. A. King, A. R. Fooks, M. Aubert, A. I. Wandeler. Paris: OIE; 2004; 47–63.
15. Poleshchuk E. M., Sidorov G. N. Comparative analysis of features of epizootiological and epidemic situation and risk of rabies infection in the Russian Federation in early XXI century. Problems of Particularly Dangerous Infections. 2020; (4): 16–25. https://doi.org/10.21055/0370-1069-2020-4-16-25 (in Russ.)
16. Gruzdev K. N., Metlin A. Ye. Animal Rabies. 2nd ed., revised and expanded. Vladimir: Federal Center for Animal Health; 2022. 442 p. (in Russ.)
17. Avilov V. M., Sochnev V. V., Savvin A. V., Goryachev I. I., Aliev A. A. Funktsionirovanie parazitarnoi sistemy beshenstva v sub”ektakh federatsii Povolzhskogo ekonomicheskogo raiona = Functioning of the rabies parasitic system in the Federal Subjects of the Volga Economic Region. Russian Journal of Veterinary Pathology. 2004; (3): 127–134. https://elibrary.ru/hsoxed (in Russ.)
18. Savchenko S. T., Chaika A. N., Romasova E. I., Maslennikova G. F., Makhonin A. A., Frolova G. I., et al. Sostoyanie epidemiologicheskoi i epizootologicheskoi situatsii v Volgogradskoi oblasti po prirodno-ochagovym i osobo opasnym infektsiyam = Epidemiological and epizootiological situation on sylvatic cycle diseases and highly dangerous infections in Volgograd Oblast. Russian Journal of Infection and Immunity. 2012; 2 (1–2): 192. https://iimmun.ru/iimm/article/view/90/89 (in Russ.)
19. Alikova G. A., Sochnev V. V., Avilov V. M., Kozyrenko O. V., Suvorin V. V., Pashkina Yu. V., et al. Rabies infection risk zones in Volgograd region. Glavnye epizootologicheskie parametry populyatsii zhivotnykh: sbornik nauchnykh trudov FGBOU VPO NGSKHA, predstavlennykh na 2-i sessii Mezhdunarodnoi nauchno-prakticheskoi konferentsii (Nizhnii Novgorod, 5–6 fevralya 2014 g.) = Key epizootiological parameters of animal populations: proceedings of the Nizhny Novgorod State Agricultural Academy presented at the 2nd Session of the International Scientific-Practical Conference (Nizhny Novgorod, 5–6 February 2014). Vol. 1. Nizhny Novgorod: BIKAR; 2015; 312–319. https:/elibrary.ru/tqijur (in Russ.)
20. Nevinskiy A. B., Arova A. A., Khlynina Yu. O. Klinicheskii sluchai beshenstva v Volgogradskoi oblasti = Clinical rabies case in Volgograd Oblast. Infektsionnye bolezni v sovremennom mire: evolyutsiya, tekushchie i budushchie ugrozy: sbornik trudov ХIII Ezhegodnogo Vserossiiskogo kongressa po infektsionnym boleznyam imeni akademika V. I. Pokrovskogo (Moskva, 24–26 maya 2021 g.) = Infectious Diseases in the Modern World: Evolution, Current and Future Threats: proceedings of the XIII Annual All-Russian Congress on Infectious Diseases named after Academician V. I. Pokrovsky (Moscow, 24–26 May 2021). Moscow: Meditsinskoe marketingovoe agentstvo; 2021; 120–121. https://elibrary.ru/jgusgf (in Russ.)
21. Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution. 2018; 4 (1):vey016. https://doi.org/10.1093/ve/vey016
22. Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018; 35 (6): 1547–1549. https://doi.org/10.1093/molbev/msy096
23. Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology. 2018; 67 (5): 901–904. https://doi.org/10.1093/sysbio/syy032
24. Sauvage T., Plouviez S., Schmidt W. E., Fredericq S. TREE2FASTA: a flexible Perl script for batch extraction of FASTA sequences from exploratory phylogenetic trees. BMC Research Notes. 2018; 11:164. https://doi.org/10.1186/s13104-018-3268-y
25. Ryabinina N. O. Nature and landscapes of Volgograd Oblast: monograph. Volgograd: Volgograd State University; 2015. 370 p. (in Russ.)
26. Kiener T. V., Zaitsev V. A. Population density and dispersal of the red fox (Vulpes vulpes L.) in the forest biom of Eastern Europe. Contemporary Problems of Ecology. 2010; 3 (1): 119–126. https://doi.org/10.1134/S1995425510010194
About the Authors
S. A. ChupinRussian Federation
Sergei A. Chupin - Cand. Sci. (Biology), Leading Researcher, Reference Laboratory for Rabies and BSE, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
E. V. Chernyshova
Russian Federation
Elena V. Chernyshova - Cand. Sci. (Veterinary Medicine), Head of Reference Laboratory for Rabies and BSE, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
R. S. Chernyshev
Russian Federation
Roman S. Chernyshev - Cand. Sci. (Biology), Junior Researcher, Reference Laboratory for African Swine Fever, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
K. N. Gruzdev
Russian Federation
Konstantin N. Gruzdev - Dr. Sci. (Biology), Professor, Chief Researcher, Information and Analysis Centre, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
A. N. Spiridonov
Russian Federation
Artem N. Spiridonov - Cand. Sci. (Veterinary Medicine), Head of Information and Analysis Centre, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
A. V. Varkentin
Russian Federation
Andrey V. Varkentin - Cand. Sci. (Veterinary Medicine), Head of Sector, Information and Analysis Center, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
N. A. Nazarov
Russian Federation
Nikolay A. Nazarov - Cand. Sci. (Biology), Leading Researcher, Reference Laboratory for Rabies and BSE, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
I. D. Zarva
Russian Federation
Ivan D. Zarva - Cand. Sci. (Medicine), Head of Department of Epidemiology, Irkutsk State Medical University.
1 Krasnogo Vosstaniya str., Irkutsk 664003
A. D. Botvinkin
Russian Federation
Aleksandr D. Botvinkin - Dr. Sci. (Medicine), Professor of Department of Epidemiology, Irkutsk State Medical University.
1 Krasnogo Vosstaniya str., Irkutsk 664003
Review
For citations:
Chupin S.A., Chernyshova E.V., Chernyshev R.S., Gruzdev K.N., Spiridonov A.N., Varkentin A.V., Nazarov N.A., Zarva I.D., Botvinkin A.D. Phylogenetic analysis of rabies virus isolates recovered from animals in Volgograd Oblast. Veterinary Science Today. 2025;14(3):241-248. https://doi.org/10.29326/2304-196X-2025-14-3-241-248



































