Scroll to:
Clostridium species diversity in cattle
https://doi.org/10.29326/2304-196X-2025-14-2-194-200
Abstract
Introduction. Clostridial infections, though relatively sporadic, are globally ubiquitous and specified by high mortality rates, resulting in substantial economic losses to agriculture. In cattle, pathogenic сlostridia cause diseases such as enterotoxemia, malignant edema, tetanus, and botulism. The most clinically significant species include Clostridium septicum, Clostridium perfringens, Clostridium chauvoei, Clostridium novyi, and Clostridium sordellii.
Objective. Study of Clostridium spp. diversity by examination of autopsy samples and sections of cattle from different regions of Russia; determination of their anatomical localization as well as antibiotic resistance of Clostridium perfringens to the most common groups of antibiotics.
Materials and methods. Throughout the study, we adhered to internationally recognized regulatory frameworks and methodological guidelines, employing standardized microbiological and mass-spectrometric methods. Antibiotic resistance was tested against multiple drug groups, such as macrolides, monobactams, penicillins, polypeptides, glycopeptides, aminoglycosides, carbapenems, lincosamides, tetracyclines, ansamycins, diaminopyrimidines, fusidic acid derivatives, etc. Clostridium isolates were recovered and identified using routine bacteriological methods coupled with MALDI-ToF mass spectrometry.
Results. Analysis of 359 biological samples resulted in isolation and identification of 137 Clostridium isolates (Paraclostridium bifermentans, Clostridium perfringens,
Clostridium tertium, Clostridium butyricum, Clostridium septicum, Clostridium sporogenes, Clostridium cadaveris, Clostridium sphenoides, Clostridium cochlearium, Clostridium sartagoforme, Clostridium chauvoei, Clostridium novyi, Clostridium sordellii, Clostridium paraputrificum, Clostridium spp.), of which 25 exhibited pathogenic potential and 17 demonstrated toxigenic properties. Сlostridia were most frequently isolated from the liver, small and large intestinal segments, and muscular tissues. Herewith, Clostridium perfringens prevailed (17.5%). This bacterium isolates demonstrated multiple drug resistance to cefixime, fusidic acid, cefotaxime, cefaclor, spectinomycin, piperacillin, clarithromycin, doripenem and doxycycline.
Conclusion. The obtained results can be used for modification of current clostridial infection treatment protocols, reformulation of immunobiological products, development of evidence-based guidelines for use of antibiotics in livestock production to mitigate antimicrobial resistance risks.
Keywords
For citations:
Shastin P.N., Savinov V.A., Laishevtsev A.I., Mandryka E.D., Fabrikantova E.A., Supova A.V. Clostridium species diversity in cattle. Veterinary Science Today. 2025;14(2):194-200. https://doi.org/10.29326/2304-196X-2025-14-2-194-200
INTRODUCTION
The genus Clostridium was first described by A. Prażmowski in 1880. Over 225 species of Clostridia have been currently identified in various regions of the planet. Clostridia are gram-positive rods that form spores. They are widespread in the environment, and are also part of the human and animal microbiome. However, only some of them are capable of causing diseases in animals [1-3]. Clostridial infections are characterized by high mortality. Due to the spore-forming ability of Clostridia, they can persist in the soil for a long time, thus posing a potential threat of the disease emergence [4-6]. The pathogen entry into the body of animals occurs mainly by ingestion of contaminated feed (alimentary route), through wounds or by inhalation. The main factors of Clostridium pathogenicity are exotoxins and enzymes [7-10], which have hemolytic, necrotizing and lethal effects. The most potent toxins of clostridial origin are botulinum and tetanus neurotoxins, as well as epsilon toxin produced by Clostridium perfringens types B and D [11-14].
The emergence of polyresistant Clostridium strains results in wider spread of clostridial infections. A number of scientists have noted low therapeutic efficacy of antibacterial drugs against the clinical manifestation of anaerobic enterotoxemia in young cattle, high mortality and the need for specific prevention [7][15-19].
According to “Galen” component of the FGIS “VetIS”, the list of registered vaccines against bovine clostridial infections in the Russian Federation is currently includes the following products: Clostrivax (Tecnovax S. A., Argentina); Coglavax (Ceva Sante Animale, France; Ceva-Phylaxia Veterinary Biologicals Company, Hungary); Clostbovac-8 (Vetbiochem LLC, Russia); Clostarm-9 (Armavir Biofactory, Russia); Cubolac (CZ Vaccines S. A. U., Spain); Antox 9 (Stavropol Biofactory, Russia); One Shot Ultra 8 (Zoetis Inc., USA); Scourguard 4KS (Zoetis Inc., USA).
The relevance and novelty of the work lies in obtaining data on the antibiotic resistance of the etiologically relevant Clostridium isolates, on the structure of the strains isolated from cattle, and on their toxigenic and pathogenic properties. The resulted data will contribute to the improvement of the clostridial infection control system in cattle, which in turn will reduce the economic losses in the livestock production.
The aim of the work was to conduct the monitoring studies to identify Clostridia, as well as to assess the level of antimicrobial resistance of Clostridium perfringens isolates recovered from cattle in various regions of Russia, and to study their toxigenic and pathogenic properties.
MATERIALS AND METHODS
The work was performed in 2022–2024 at the Laboratory for Diagnostics and Control of Antibiotic Resistance of Pathogens of the Most Clinically Significant Infectious Animal Diseases of the Federal Scientific Centre VIEV, as part of the state project (FGUG-2025-0003) supported by the Ministry of Science and Higher Education of the Russian Federation. As a result of our own research, monitoring data was obtained and the practical part was completed. Sectional and autopsy materials collected from cattle were delivered from various regions of Russia: Nizhny Novgorod, Moscow, Leningrad, Ryazan, Novosibirsk, Penza Oblasts and Republic of Mordovia.
Biological material. A total of 359 samples were examined (liver, heart, spleen, lung, kidney, muscle, small and large intestines, stomach, hoof sections, amniotic fluid, etc.).
Recovery of isolates, determination of their pathogenic and toxigenic properties. The study aimed at the recovery of the isolates of the microorganisms that are etiologically most relevant for commercial animal husbandry, namely the Clostridiaceae family, was implemented in accordance with GOST 26503-85 “Agricultural animals. Methods for laboratory diagnostics of clostridium”1.
Identification of Clostridia. Species identification of the microorganisms was performed by mass spectrometry using MALDI Biotyper system (Bruker Daltonik GmbH, Germany) according to the “Guidelines for the identification of microorganisms using MALDI Biotyper mass spectrometer for the examination of food raw materials and food products” (approved by the Rosselkhoznadzor RTC on 3 April 2014).
Antibiotic resistance of the microbial cultures was determined by disc diffusion method in accordance with Methodological Guidelines MUK 4.2.1890-04 “Guidelines for susceptibility testing of microorganisms to antibacterial agents”2. Within the research activities, antibacterial drugs of various groups were used (HiMedia Laboratories Pvt Ltd., India): macrolides (azithromycin 15 µg, clarithromycin 15 µg, pristinamycin 15 µg, spiramycin 30 µg, tylosin 15 µg, erythromycin 15 µg), monobactams (aztreonam 30 µg), penicillins (amoxiclav 30 µg, amoxicillin 25 µg, ampicillin 25 µg, benzylpenicillin 10 µg, carbenicillin 100 µg, piperacillin 100 µg), polypeptides (bacitracin 10 µg, polymyxin B 50 µg), chloramphenicol 30 µg, glycopeptides (vancomycin 30 µg), aminoglycosides (gentamicin 30 µg, kanamycin 30 µg, spectinomycin 100 µg, streptomycin 25 µg), carbapenems (doripenem 10 µg), lincosamides (clindamycin 2 µg, lincomycin 10 µg), fluoroquinolones (levofloxacin 5 µg, norfloxacin 10 µg, ofloxacin 5 µg, pefloxacin 5 µg, ciprofloxacin 30 µg, enrofloxacin 10 µg), tetracyclines (oxytetracycline 30 µg, tetracycline 30 µg, chlortetracycline 30 µg, doxycycline 30 µg), ansamycins (rifampicin 15 µg), sulfonamides (sulfadiazine 100 µg, sulfafurazole 300 µg), diaminopyrimidines (trimethoprim 25 µg), cephalosporins (cefixime 5 µg, cefazolin 30 µg, cefaclor 30 µg, cefalexin 30 µg, cefotaxime 30 µg, cefepime 30 µg, cefoperazone 75 µg, cefpirome 30 µg, ceftriaxone 30 µg), phosphonic acid derivatives (fosphomycin 50 µg), fusidines (fusidic acid 30 µg).
The results were interpreted in accordance with CLSI (Clinical and Laboratory Standards Institute) and EUCAST (European Committee on Antimicrobial Susceptibility Testing) recommendations [20][21].
The results were statistically processed using Microsoft Excel.
RESULTS AND DISCUSSION
As a result of the studies, 137 Clostridium isolates were recovered and identified: Paraclostridium bifermentans, Clostridium perfringens, Clostridium tertium, Clostridium butyricum, Clostridium septicum, Clostridium sporogenes, Clostridium cadaveris, Clostridium sphenoides, Clostridium cochlearium, Clostridium sartagoforme, Clostridium chauvoei, Clostridium novyi, Clostridium sordellii, Clostridium paraputrificum, Clostridium spp.
Diversity of Clostridium spp. circulating in the Russian Federation has been established, which is shown in Figure 1.
Prevalence of C. perfringens was established – 17.5%, followed by C. tertium – 13.1%, C. sordellii – 11.7%, C. chauvoei – 11.0%, C. novyi – 9.5%, P. bifermentans and C. butyricum – 8.0%, C. sporogenes – 6.6%, Clostridium spp. – 5.0%, C. septicum – 3.0%, C. cadaveris – 2.2%, C. sphenoides and C. cochlearium – 1.5% each, the smallest proportion is made up of C. sartagoforme and C. paraputrificum isolates – 0.7%.
Results of determination of antibiotic resistance of C. perfringens isolates (n = 24) recovered from cattle in various regions of the Russian Federation are demonstrated in Figure 2.
According to the obtained data, it can be concluded that C. perfringens isolates (n = 24) demonstrated resistance to cefixime, fusidic acid, cefotaxime, cefaclor, spectinomycin, piperacillin, clarithromycin, doripenem, and doxycycline. Antibiotic resistance to ampicillin demonstrated 85% of the isolates, to amoxicillin, chlortetracycline, vancomycin, rifampicin and ciprofloxacin – 80%, to tylosin and amoxiclav – 75%, to sulfadiazine, cefalexin, ofloxacin and polymyxin B – 60%, to pefloxacin and cefoperazone – 55%, to benzylpenicillin, clindamycin, ceftriaxone and chloramphenicol – 50%, to enrofloxacin, cefazolin, tetracycline and streptomycin – 45% of the isolates; 40% of C. perfringens isolates were resistant to bacitracin, norfloxacin, fosfomycin; 35% of the isolates demonstrated resistance to levofloxacin, lincomycin, oxytetracycline; 25% of isolates were resistant to erythromycin, spiramycin and gentamicin and 20% – to azithromycin, cefepime and cefpirome. All studied C. perfringens isolates were susceptible to sulfafurazole and carbenicillin (100%), trimethoprim – 90%, azithromycin – 70%, levofloxacin – 65%, and kanamycin – 45% of the isolates. All the tested strains were intermediately susceptible to aztreonam and pristinamycin, 75% of the isolates – to spiramycin, 60% – to fosfomycin, and 55% – to cefazolin, 45% – to kanamycin.
Among 137 recovered Clostridium isolates, 25 demonstrated pathogenic properties and 17 had toxigenic properties. The obtained data is presented graphically as a percentage in Figures 3 and 4.
In most cases, C. perfringens isolates possessed pathogenic properties (6.6%). Pathogenicity factors were detected in 5.1% of C. novyi strains, 4.4% of C. chauvoei isolates, in 1.5% of C. septicum strains and in 0.7% of Clostridium spp. isolates. Toxigenic properties were determined for C. sordellii (3.7%), C. perfringens (3.7%), C. novyi (3.0%), C. septicum (1.5%) and Clostridium spp. (0.7%).
The localization sites of Clostridia in cattle are presented in the Table.
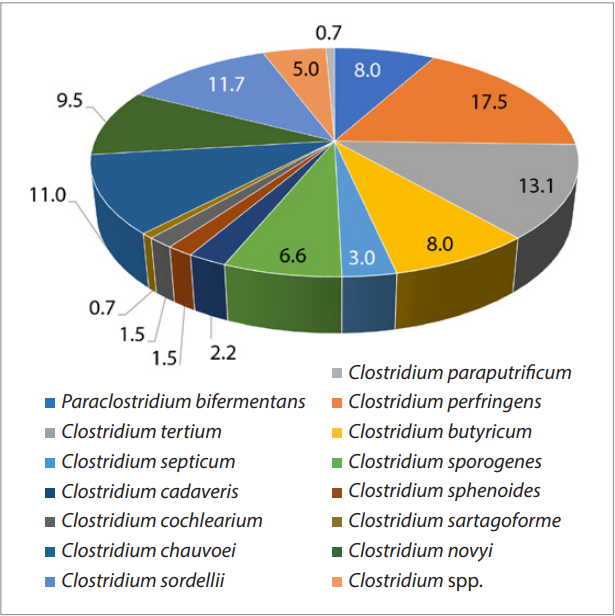
Fig. 1. Species diversity of Clostridium isolates circulating in the Russian Federation (n = 137), %
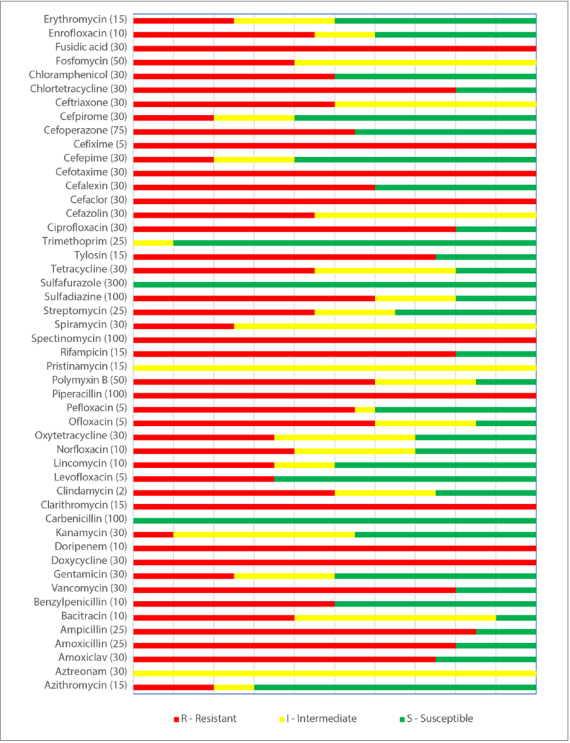
Fig. 2. Antibiotic resistance of C. perfringens isolates (n = 24) recovered from cattle
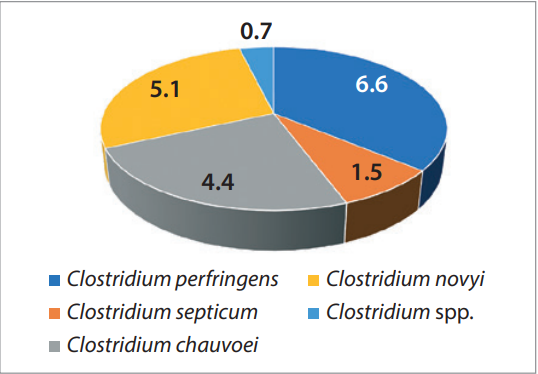
Fig. 3. Species composition of Clostridium isolates with pathogenic properties, %
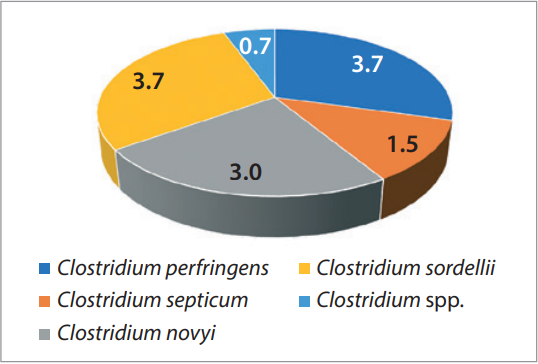
Fig. 4. Species composition of Clostridium isolates with toxic properties, %
Table
Localization of Clostridia in cattle
|
Clostridium species |
Biological material |
||||||||||
|
Heart |
Liver |
Spleen |
Lung |
Kidney |
Muscle |
Small intestine |
Large intestine |
Stomach |
Hoof sections |
Amniotic fluid |
|
|
Paraclostridium bifermentans |
– |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
+ |
– |
|
Clostridium tertium |
– |
+ |
+ |
– |
– |
+ |
+ |
+ |
– |
– |
– |
|
Clostridium perfringens |
– |
+ |
+ |
– |
+ |
+ |
+ |
+ |
+ |
– |
+ |
|
Clostridium butyricum |
– |
+ |
– |
– |
– |
– |
+ |
+ |
+ |
– |
– |
|
Clostridium cochlearium |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
|
Clostridium sartagoforme |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
|
Clostridium septicum |
– |
+ |
– |
– |
+ |
+ |
– |
+ |
– |
– |
– |
|
Clostridium sporogenes |
– |
+ |
– |
– |
– |
+ |
+ |
+ |
– |
– |
– |
|
Clostridium sphenoides |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
|
Clostridium chauvoei |
– |
+ |
– |
– |
– |
– |
+ |
+ |
+ |
– |
– |
|
Clostridium novyi |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
Clostridium sordellii |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
Clostridium paraputrificum |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
Clostridium spp. |
– |
+ |
– |
– |
– |
+ |
+ |
+ |
– |
– |
– |
|
Clostridium cadaveris |
– |
+ |
– |
– |
+ |
– |
– |
+ |
– |
– |
– |
According to the demonstrated data, Clostridia were most often isolated from the liver, small and large intestine and from muscles.
Clostridia are widespread bacteria that cause diseases in animals, birds and humans. Antibiotic resistance is a serious challenge for the veterinary medicine due to the fact that 80% of all antibiotics in the world are used in agriculture, inter alia as feed additives and growth promoters. The results obtained during the present study on antibiotic resistance to cefotaxime are consistent with the data reported by N. A. Bezborodova et al. [7], H. A. Ahmed et al. [22]. In the studies carried out by the Iranian researchers F. Khademi et al., resistance of C. perfringens to ampicillin (25.8%), erythromycin (32.9%), gentamicin (45.4%), tetracycline (19.5%), amoxicillin (19.3%), bacitracin (89.1%) was reported [23]. A group of scientists from China and Pakistan studied eleven of the most commonly used antibiotics, two of them had no inhibitory effect, five were effective, and four had moderate effect against C. perfringens. Lincomycin and amikacin did not inhibit the isolates, tetracycline, penicillin, erythromycin and oxytetracycline inhibited Clostridium growth to a lesser extent. The scientists have concluded that it was advisable to use several types of antibiotics, which was a more effective approach to inhibit the bacterial infection [24]. Researchers from Ivory Coast determined in their studies that the level of antibiotic resistance of C. perfringens to tetracycline, doxycycline, chloramphenicol, and erythromycin ranged from 20 to 50% [25]. A group of scientists from South Korea, when studying the prevalence and resistance of C. perfringens to antibiotics, found that resistance to tetracycline was 100%, to ampicillin – 31.6%, to chloramphenicol – 68.4%, to metronidazole – 34.2% and to imipenem – 71%. The researchers also noted an important point of the combined resistance of 78.9% of the isolates to several antimicrobial drugs [26].
CONCLUSION
As a result of the examination of the sections and pathological materials from cattle in 2022–2024, 137 Clostridium isolates were recovered, of which 25 demonstrated pathogenic properties, and 17 – toxigenic ones. The most common Clostridium localization sites included liver, large and small intestine, muscles, and stomach. The bacteria were also detected in kidneys, spleen, amniotic fluid, and hoof swabs.
Monitoring studies aimed at the determination of the antimicrobial resistance of C. perfringens isolates revealed their resistance to cefixime, fusidic acid, cefotaxime, cefaclor, spectinomycin, piperacillin, clarithromycin, doripenem, and doxycycline.
The results of this study can be used to modify existing treatment protocols for clostridial infections, adjust the composition of immunobiological products, and develop recommendations for the use of antibiotics in animal husbandry to reduce the risk of antimicrobial resistance developing.
1. https://base.garant.ru/5916932 (in Russ.)
2. https://docs.cntd.ru/document/1200038583 (in Russ.)
References
1. Kapustin A. V., Aliper T. I. Epizootologiya i profilaktika klostridiozov krupnogo rogatogo skota = Epizootology and prevention of bovine clostridial diseases. Edinyi mir – edinoe zdorov’e: materialy VII Mezhdunarodnogo veterinarnogo kongressa (Ufa, 19–21 aprelya 2017 g.) = One World – One Health: Materials of VII International Veterinary Congress (Ufa, April 19–21, 2017). Moscow: Russian Veterinary Association; 2017; 106–108. https://elibrary.ru/zarmnr (in Russ.)
2. Sudorgina T. E., Glotova T. I., Nefedchenko A. V., Koteneva S. V., Velker D. A., Glotov A. G. The frequency of bacterial isolation of Clostri dium spp. and their associations in various forms of clostridiosis in cattle. Siberian Herald of Agricultural Science. 2024; 54 (3): 55–62. https://doi.org/10.26898/0370-8799-2024-3-6
3. Salvarani F. M., Vieira E. V. Clostridial infections in cattle: a comprehensive review with emphasis on current data gaps in Brazil. Animals. 2024; 14 (20):2919. https://doi.org/10.3390/ani14202919
4. Robi D. T., Mossie T., Temteme S. A comprehensive review of the common bacterial infections in dairy calves and advanced strategies for health management. Veterinary Medicine: Research and Reports. 2024; 15: 1–14. https://doi.org/10.2147/vmrr.s452925
5. Santos B. L., Ladeira S. R. L., Riet-Correa F., Soares M. P., Marcolongo- Pereira C., Sallis E. S. V., et al. Clostridial diseases diagnosed in cattle from the South of Rio Grande do Sul, Brazil. A forty-year survey (1978–2018) and a brief review of the literature. Pesquisa Veterinária Brasileira. 2019; 39 (7): 435–446. http://doi.org/10.1590/1678-5150-pvb-6333
6. Popoff M. R., Bouvet P. Clostridial toxins. Future Microbiology. 2009; 4 (8): 1021–1064. https://doi.org/10.2217/fmb.09.72
7. Bezborodova N. A., Sokolova O. V., Kozhukhovskaya V. V., Tomskikh O. G., Pechura E. V., Suzdal’tseva M. A. Pathogenic species of Clostridia and their antibiotic resistance, virulence factors, and genomic features. Innovations and Food Safety. 2023; (3): 39–51. https://doi.org/10.31677/23110651-2023-41-3-39-51 (in Russ.)
8. Glotova T. I., Terentyeva T. E., Glotov A. G. Pathogens and age susceptibility of cattle to clostridiosis. Siberian Herald of Agricultural Science. 2017; 47 (1): 90–96. https://elibrary.ru/ylkevt (in Russ.)
9. Kapustin A. V., Laishevtcev A. I., Ivanov E. V., Danilyuk A. V. Species diversity of Clostridia causing malignant edema in cattle. IOP Conference Series: Earth and Environmental Science. 2020; 548 (7):072041. https://doi.org/10.1088/1755-1315/548/7/072041
10. Meltsov I. V., Blokhin A. A., Sukhinin A. A., Batomunkuyev A. S., Kutuzova L. A. Epizootological investigation of an outbreak of emphysematous carbuncle in cattle (case study in the Irkutsk Region). International Bulletin of Veterinary Medicine. 2024; (3): 84–94. https://doi.org/10.52419/issn2072-2419.2024.3.84 (in Russ.)
11. Rings D. M. Clostridial disease associated with neurologic signs: tetanus, botulism, and enterotoxemia. Veterinary Clinics of North America: Food Animal Practice. 2004; 20 (2): 379–391. https://doi.org/10.1016/j.cvfa.2004.02.006
12. Otter A., Uzal F. A. Clostridial diseases in farm animals: 2. Histo toxic and neurotoxic diseases. In Practice. 2020; 42 (5): 279–288. https://doi.org/10.1136/inp.m1984
13. Otter A., Uzal F. A. Clostridial diseases in farm animals: 1. Enterotoxaemias and other alimentary tract infections. In Practice. 2020; 42 (4): 219–232. https://doi.org/10.1136/inp.m1462
14. Jing W., Pilato J. L., Kay C., Feng S., Tuipulotu D. E., Mathur A., et al. Clostridium septicum α-toxin activates the NLRP3 inflammasome by engaging GPI-anchored proteins. Science Immunology. 2022; 7 (71):eabm1803. https://doi.org/10.1126/sciimmunol.abm1803
15. Kapustin A. V., Laishevtsev A. I., Motorygin A. V. Emphysematous carbuncle in cattle. Russian Journal of Agricultural and SocioEconomic Sciences. 2021; (1): 149–156. https://doi.org/10.18551/rjoas.2021-01.20
16. Kapustin A. V. Development of a vaccine against blackleg of cattle. Russian Journal of Agricultural and SocioEconomic Sciences. 2016; (5): 97–102. https://doi.org/10.18551/rjoas.2016-05.13 (in Russ.)
17. Terentjeva T. E., Glotova T. I., Koteneva S. V., Glotov A. G. Species spectrum of bacteria of genus Clostridium isolated from cattle on big dairy farms. Russian Veterinary Journal. Productive Animals. 2016; (1): 5–8. https://elibrary.ru/vmdwdn (in Russ.)
18. Kozlova A. D., Gorbacheva N. S., Klimenkova O. V., Laishevtcev A. I., Kapustin A. V., Yatsentyuk S. P. The use of molecular genetic techniques for the typing of Clostridium perfringens. Russian Journal of Agricultural and SocioEconomic Sciences. 2017; (3): 188–194. https://doi.org/10.18551/rjoas.2017-03.23 (in Russ.)
19. Sklyarov O. D., Kapustin A. V., Laishevtcev A. I., Gulyukin A. M. Interference of components in a polyvalent vaccine against clostridiosis of cattle and small cattle. Russian Veterinary Journal. 2017; (1): 20–23. https://elibrary.ru/xxyqvr (in Russ.)
20. Clinical and Laboratory Standards Institute (CLSI). https://clsi.org
21. European Committee on Antimicrobial Susceptibility Testing ( EUCAST). https://www.eucast.org
22. Ahmed H. A., El Bayomi R. M., Hamed R. I., Mohsen R. A., El- Gohary F. A., Hefny A. A., et al. Genetic relatedness, antibiotic resistance, and effect of silver nanoparticle on biofilm formation by Clostridium perfringens isolated from chickens, pigeons, camels, and human consumers. Veterinary Sciences. 2022; 9 (3):109. https://doi.org/10.3390/vetsci9030109
23. Khademi F., Sahebkar A. The prevalence of antibiotic-resistant Clostridium species in Iran: a meta-analysis. Pathogens and Global Health. 2019; 113 (2): 58–66. https://doi.org/10.1080/20477724.2019.1603003
24. Khan M. U. Z., Humza M., Yang S., Iqbal M. Z., Xu X., Cai J. Evaluation and optimization of antibiotics resistance profile against Clostridium perfringens from buffalo and cattle in Pakistan. Antibiotics. 2021; 10 (1):59. https://doi.org/10.3390/antibiotics10010059
25. Kouassi K. A., Dadie A. T., N’Guessan K. F., Dje K. M., Loukou Y. G. Clostridium perfringens and Clostridium difficile in cooked beef sold in Côte d’Ivoire and their antimicrobial susceptibility. Anaerobe. 2014; 28: 90–94. https://doi.org/10.1016/j.anaerobe.2014.05.012
26. Jang Y.-S., Kim D.-H., Bae D., Kim S.-H., Kim H., Moon J.-S., et al. Prevalence, toxin-typing, and antimicrobial susceptibility of Clostridium perfringens from retail meats in Seoul, Korea. Anaerobe. 2020; 64:102235. https://doi.org/10.1016/j.anaerobe.2020.102235
About the Authors
P. N. ShastinRussian Federation
Pavel N. Shastin, Cand. Sci. (Veterinary Medicine), Senior Researcher, Laboratory for Diagnostics and Control of Antibiotic Resistance of Pathogens of the Most Clinically Significant Infectious Animal Diseases,
24/1, Ryazansky prospekt, Moscow 109428.
V. A. Savinov
Russian Federation
Vasiliy A. Savinov, Cand. Sci. (Biology), Senior Researcher, Laboratories of Mycology and Antibiotics named after A. H. Sarkisov,
24/1, Ryazansky prospekt, Moscow 109428.
A. I. Laishevtsev
Russian Federation
Aleksey I. Laishevtsev, Cand. Sci. (Biology), Leading Researcher, Acting Head, Laboratory for Diagnostics and Control of Antibiotic Resistance of Pathogens of the Most Clinically Significant Infectious Animal Diseases,
24/1, Ryazansky prospekt, Moscow 109428.
E. D. Mandryka
Russian Federation
Ekaterina D. Mandryka, Microbiologist, Laboratory for Diagnostics and Control of Antibiotic Resistance of Pathogens of the Most Clinically Significant Infectious Animal Diseases,
24/1, Ryazansky prospekt, Moscow 109428.
E. A. Fabrikantova
Russian Federation
Elizaveta A. Fabrikantova, Microbiologist, Laboratory for Diagnostics and Control of Antibiotic Resistance of Pathogens of the Most Clinically Significant Infectious Animal Diseases,
24/1, Ryazansky prospekt, Moscow 109428.
A. V. Supova
Russian Federation
Anastasia V. Supova, Researcher, Laboratory for Diagnostics and Control of Antibiotic Resistance of Pathogens of the Most Clinically Significant Infectious Animal Diseases,
24/1, Ryazansky prospekt, Moscow 109428.
Review
For citations:
Shastin P.N., Savinov V.A., Laishevtsev A.I., Mandryka E.D., Fabrikantova E.A., Supova A.V. Clostridium species diversity in cattle. Veterinary Science Today. 2025;14(2):194-200. https://doi.org/10.29326/2304-196X-2025-14-2-194-200



































