Scroll to:
Development of test-system for detection of H5 and H7 avian influenza virus RNA by multiplex real-time RT-PCR assay using internal control
https://doi.org/10.29326/2304-196X-2025-14-1-40-46
Abstract
Introduction. High pathogenicity avian influenza is a dangerous highly contagious viral infection of domestic and wild birds that recently has become widespread in Europe, Asia, Africa and Americas. The causative agent of the disease is type A influenza virus of subtypes H5 and H7. Real-time RT-PCR is one of the most rapid and effective techniques for avian influenza virus identification and typing, so development of the test system based on this technique with internal control to be used for control of the reaction main stages is of current importance. At the same time, the multiplex format of RT-PCR allows for simultaneous identification of several targets that reduces the consumption of reagents and the reaction time.
Objective. Development of test-system for detection of H5 and H7 avian influenza virus RNA with multiplex real-time RT-PCR in biological samples and its characterization.
Materials and methods. H5, H7, H3, H4, H10, H16 avian influenza virus, Newcastle disease virus, infectious bursal disease virus, infectious bronchitis virus, Marek’s disease virus, avian adenovirus isolates were used. MS2 bacteriophage was used as internal control.
Results. Optimal primer-probe combinations were selected, test-system characteristics were determined: specificity for homologous and heterologous avian disease viruses, analytical sensitivity, reaction amplification efficiency, repeatability and reproducibility.
Conclusion. Determination of the developed test system validation parameters has shown that it is specific only for H5 and H7 avian influenza virus, its analytical sensitivity for each subtype was 1.5 lg EID50/cm3 , and the amplification efficiency was 92 and 97%, respectively. The test system was validated through its use for testing biological samples submitted to the laboratory, the test results were consistent with the results of tests with standard diagnostic methods used in the Reference Laboratory for Avian Viral Diseases of the Federal Centre for Animal Health.
Keywords
For citations:
Grekhneva A.D., Zinyakov N.G., Andriyasov A.V., Kozlov A.A., Ovchinnikova E.V., Andreychuk D.B., Zhestkov P.D., Chvala I.A. Development of test-system for detection of H5 and H7 avian influenza virus RNA by multiplex real-time RT-PCR assay using internal control. Veterinary Science Today. 2025;14(1):40-46. https://doi.org/10.29326/2304-196X-2025-14-1-40-46
INTRODUCTION
Avian influenza is one of the most dangerous viral diseases of poultry and wild birds, that affects primarily respiratory and digestive systems. The disease is caused by virus of genus Alphainfluenzavirus, Orthomyxoviridae family. The virus genome is a single-stranded (–)RNA composed of the 8 segments allowing high-rate virus evolution owing to reassortment [1]. Avian influenza virus (AIV) is classified into 16 subtypes by hemagglutinin (HA) and 9 subtypes by neuraminidase based on antigenic differences in surface proteins [2].
The natural reservoir of the AIV is wild waterfowl. The infected wild waterfowl are asymptomatic or demonstrate mild clinical signs. The virus naturally spread along the migration routes of wild migratory birds and at the same time is transmitted to poultry [3][4]. High pathogenicity AIV is the most dangerous for poultry industry, since it can cause severe, rapidly developing disease with 100% mortality. High pathogenicity AIVs are believed to evolve under natural conditions from low-pathogenic H5 and H7 viruses through point mutations in HA gene causing accumulation of multiple basic amino acid at the HA cleavage site [4][5][6][7][8]. High pathogenicity avian influenza is to be notified to the World Organization for Animal Health (WOAH), regardless of the causative virus subtype.
Outbreaks of the disease caused by H5 high pathogenicity AIV were regularly reported in poultry and wild birds in the Russian Federation in 2021 – early 2024 (H5N1, H5N5 and H5N8 virus isolates were recovered) [9]. In 2024, high pathogenicity avian influenza outbreak caused by H7N3 AIV was reported in poultry kept at a poultry establishment in Australia [10]. Previously, disease cases, including cases in humans, caused by H7 AIV were reported in the North and South American, European, African and Asian countries [11][12].
Current high pathogenicity avian influenza situation in the Russian Federation requires ongoing monitoring. Real-time reverse transcription-polymerase chain reaction (real-time RT-PCR) is the most rapid and accurate technique for detection of AIV RNA in biological materials from various poultry and wild bird species that enables simultaneous virus typing. This technique has high sensitivity, specificity, and is relatively rapid allowing high throughput.
Highly effective test system for detection of H5 and H7 AIV RNA by real-time RT-PCR is proposed in this paper. The developed test system contains exogenous internal control, that allows control of the main reaction stages (extraction of nucleic acids, reverse transcription and PCR) and elimination of false negative results.
The study was aimed at the development of real-time RT-PCR-based test system enabling simultaneous detection H5 and H7 virus RNAs in biological materials and control of the reaction procedure at all stages, starting with the nucleic acid extraction.
MATERIALS AND METHODS
Viruses. Isolates of AIV of various subtypes, Newcastle disease, infectious bursal disease, infectious bronchitis, Marek’s disease viruses and avian adenovirus obtained from the working collection of the Reference Laboratory for Avian Viral Diseases of the Federal Centre for Animal Health were used (Table 1). MS2 bacteriophage with an infectious activity titre of 10⁶ PFU/cm³ served as internal control [13].
Table 1
Avian virus isolates used for the study
|
Virus |
Strain/isolate |
|
H5N2 avian influenza virus |
A/duck/Italy/5952/2015 |
|
H5N2 avian influenza virus |
A/avian/Italy/6558/2015 |
|
H5N2 avian influenza virus |
A/duck/Italy/6926/2017 |
|
H5N1 avian influenza virus |
A/duck/Altai/469/2014 |
|
H5N1 avian influenza virus |
A/dalmatian pelican/Astrakhan/485-1/2022 |
|
H5N5 avian influenza virus |
A/shelduck/Kalmykia/1814-1/2021 |
|
H5N8 avian influenza virus |
A/duck/KChR/1590-20/2020 |
|
H7N2 avian influenza virus |
A/chicken/Italy/1670/2015 |
|
H7N3 avian influenza virus |
A/turkey/Italy/9289/02 |
|
H7N7 avian influenza virus |
A/duck/Italy/4932/2018 |
|
H3N8 avian influenza virus |
A/wild duck/Primorsky/1872-13/21 |
|
H4N6 avian influenza virus |
A/wild duck/Primorsky/1872-11/21 |
|
H9N2 avian influenza virus |
A/chicken/Udmurtya/2008-1/21 |
|
H9N2 avian influenza virus |
A/gull/Tyva/767-113/21 |
|
H10N7 avian influenza virus |
A/wild duck/Primorsky/1872-13/21 |
|
H16N3 avian influenza virus |
A/mallard/Khabarovsk/12/14 |
|
Infectious bronchitis virus |
H-120 |
|
Infectious bursal disease virus |
Winterfield 2512 |
|
Newcastle disease virus |
LaSota (genotype II) |
|
Avian adenovirus |
KR95 (type C) |
|
Marek’s disease virus |
3004 |
RNA extraction was performed using a “RIBO-prep” reagent kit for RNA/DNA extraction from clinical samples (AmpliSens®, Russia) according to the manufacturer instructions. At the extraction stage, internal control was added to each sample (including negative extraction control), 0.01 mL of internal control per sample.
Primers and probes. Several sets of primers and probes for amplification of H5 and H7 AIV HA gene fragments were selected and tested based on the analysis of publications on development of test systems and methods for detection of H5/H7 AIV in biological samples with real-time RT-PCR [14][15][16][17]. Since real-time RT-PCR was performed in a multiplex format, the dyes included in the TaqMan probes were selected in such a way as to generate a stable fluorescent signal and not inhibit the signals in other detection channels (Green/H5, Orange/H7, Crimson/internal control). The selected primers and probes were synthesized by the Syntol company (Russia), and the specific primers and probe for internal control [13] were synthesized by the Alkor Bio Company Ltd. (Russia).
Real-time RT-PCR was performed in one step using amplification reagents manufactured by the Syntol company (Russia) in Rotor-Gene 6000 programmable amplifier (Corbett Research Pty Ltd, Australia). The reaction mix (20 µL per sample) contained: deionized (bidistilled) water – 5.35 µL; 10× PCR buffer – 2.5 µL; 25 mM MgCl2 solution – 4 µL; 25 mM deoxynucleoside triphosphate (dNTP) solution – 0.4 µL; forward and reverse primer solutions for AIV/H5, 10 pmol/µL – 1 µL of each primer; fluorescent probe for AIV/H5, 10 pmol/µL – 0.75 µL; forward and reverse primer solutions for AIV/H7, 10 pmol/µL – 1 µL of each primer; fluorescent probe for AIV/H7, 10 pmol/µL – 0.75 µL; forward and reverse primer solutions, fluorescent probe solution for MS2, 10 pmol/µL – 0.5 µL of each primer; SynTaq DNA polymerase – 0.25 µL; MMLV-revertase – 0.5 µL. The reaction was carried out according to the following procedure: reverse transcription – 20 min at 40 °C; polymerase activation – 8 min at 95 °C; 40 PCR cycles – 10 s at 95 °C; 35 s at 55 °C; 15 s at 72 °C. The fluorescence signal was detected at the stage of primer annealing using the Green/H5, Orange/H7, and Crimson/internal control channels.
The test system was examined for its specificity by performing real-time RT-PCR using RNAs extracted from homologous and heterologous viruses (Table 1).
The test system was assessed for its analytical sensitivity by performing real-time RT-PCR with extracted RNA of serial 10-fold dilutions (10⁻⁸–10⁻³) of a virus-containing suspension (AIV strains: A/duck/KChR/1590-20/2020 H5N8 and A/turkey/Italy/9289/02 H7N3, initial infectivity titre was 8.5 lg EID50/cm³) with internal control in triplicate for each dilution. The reaction sensitivity for each sample was estimated as the virus amount (measuring units – lg EID50/cm³) corresponding to the last dilution, at which at least 95% of positive results were obtained (in 20 repeats) [18].
For repeatability assessment, positive samples were tested 3 times in 5 repeats during three days. Mean threshold cycle (Ct), standard deviation, and coefficient of variation for the obtained results were determined within one real-time RT-PCR run and between runs.
To determine the reaction efficiency (E), the results obtained during the reaction runs for analytical sensitivity testing were used. The reaction efficiency was estimated after plotting a linear regression graph (in the coordinates “virus dilution” / “threshold amplification cycle Ct”) according to the following formula:
E = (10(⁻¹/m) – 1) × 100%,
where m is the slope coefficient of the straight line [19][20][21].
RESULTS AND DISCUSSION
Optimal combinations of primers and probes were determined based on the results of testing of primer and probe combinations for amplification of H5 and H7 AIV HA gene fragments. Nucleotide sequences are shown in Table 2.
Table 2
Primers and probes used for amplification of H5 and H7 AIV HA gene fragments
|
Name |
Oligonucleotide structure |
|
H5LH1 |
ACATATGACTACCCACARTATTCAG |
|
H5RH1 |
AGACCAGCTAYCATGATTGC |
|
H5Zond |
(FAM)TCWACAGTGGCGAGTTCCCTAGCA(RTQ1) |
|
LH6H7 |
GGCCAGTATTAGAAACAACACCTATGA |
|
RH4H7 |
GCCCCGAAGCTAAACCAAAGTAT |
|
H7Zond |
(ROX)CCGCTGCTTAGTTTGACTGGGTCAATCT(BHQ2) |
Selected sets were tested in real-time RT-PCR with AIV strains of H5N2, H5N1, H5N5, H5N8, H7N2, H7N3 and H7N7 subtypes and internal control in mono- and multiplex reaction format. Threshold cycle value, above which the reaction results should be considered negative, was set at 36.00 for the Green/H5 and Orange/H7 channels. In all tests, H5 or H7 AIV RNAs were confirmed to be present only in the samples containing the relevant virus subtype.
Optimal internal control concentration was determined by performing real-time RT-PCR using several 10-fold dilutions of the MS2 virus-containing suspension (10⁵–10⁷ PFU/cm³) and simultaneous identification of one or two specific targets of the test system (AIV/H5 and AIV/H7). Based on the test results, concentration of 10⁶ PFU/cm³ was selected that enabled a stable fluorescent signal increase in detection channel for internal control without inhibiting the signal in other channels for AIV/H5 and AIV/H7 (Fig. 1) and did not exceed sensitivity of the primers-probe system for internal control while simultaneously identifying AIV/H5 and AIV/H7 at high concentrations. When the Ct value is > 35 for Crimson/internal control detection channel, the results of the entire study are considered unreliable, i.e. errors were made at the stage of nucleic acid extraction, reverse transcription, or PCR, or the test sample contains impurities capable of inhibiting the reaction.
Test of real-time RT-PCR test-system for its specificity using extracted RNAs of H3, H4, H9, H10, H16 AIV and other RNA- and DNA-containing viruses (Newcastle disease, infectious bursal disease, infectious bronchitis, Marek’s disease viruses, adenovirus) showed the absence of cross-reactions with the above-listed pathogens.
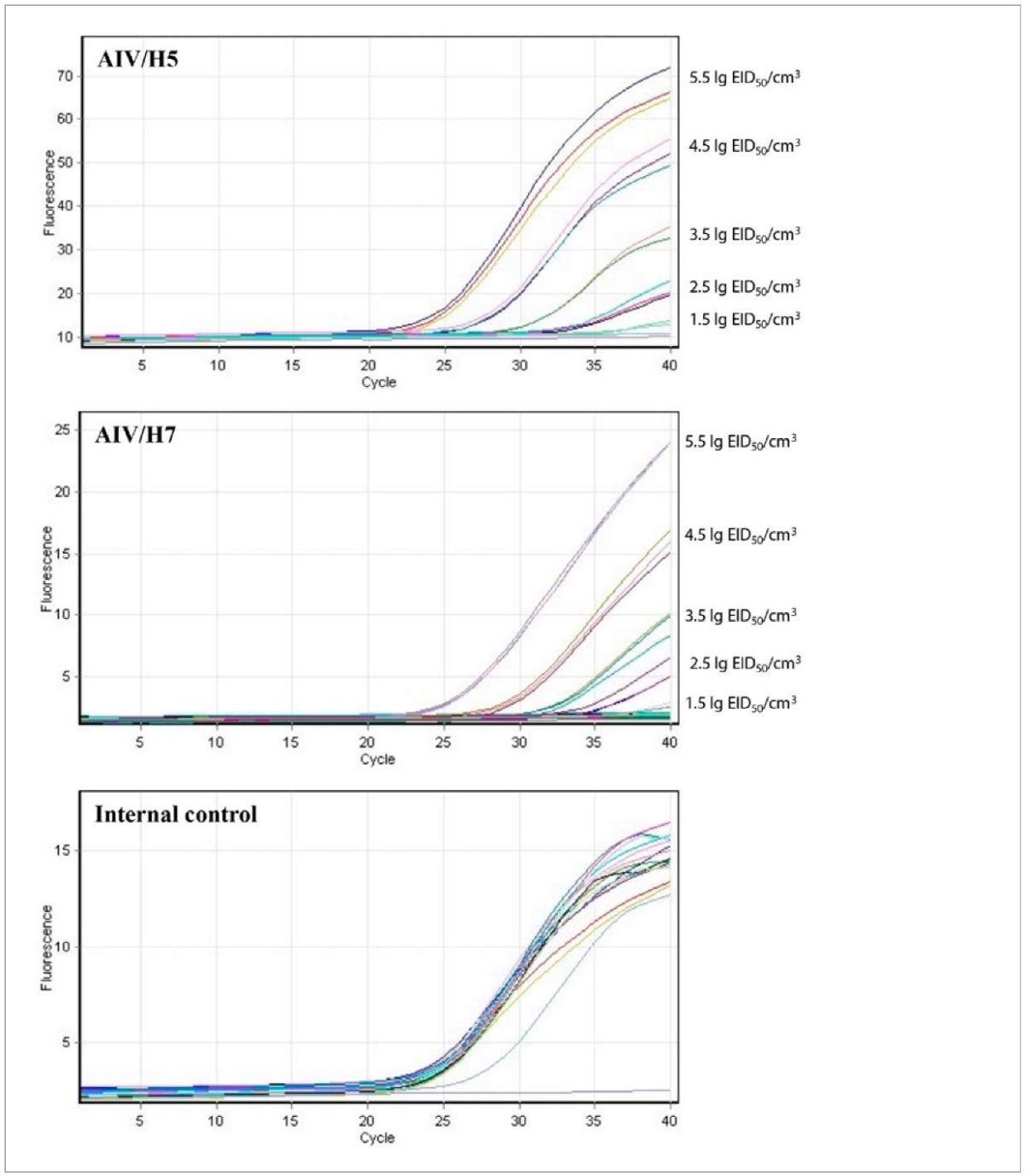
Fig. 1. Graphs of fluorescence intensity increase during real-time RT-PCR on Green (10-fold AIV/H5 dilutions), Orange (10-fold AIV/H7 dilutions) and Crimson (internal control) channels
Analytical sensitivity of the test-system used for testing 10-fold dilutions of the virus with an infectious titre of 8.5 lg EID50/cm³ (Fig. 1, Table 3) for AIV/H5 the sensitivity limit corresponded to 10⁻⁷ dilution of the virus (in 20 repeats a positive result was obtained in 95% of cases) at the average Ct value of 34.16 ± 0.54 and a coefficient of variation of 1.59%; for AIV/H7 the sensitivity limit corresponded to a virus dilution of 10⁻⁷ (in 20 repeats, positive result was obtained in 100% of cases) with average Ct value of 35.17 ± 0.65 and a coefficient of variation of 1.84%.
Table 3
Real-time RT-PCR Ct values for 10-fold AIV/Н5 and AIV/Н7 dilutions
|
Detection channel / AIV subtype (virus titre in the initial suspension was 8.5 lg ED50/cm³) |
The average Ct value for dilution, n = 3 |
|||||
|
10⁻³ |
10⁻⁴ |
10⁻⁵ |
10⁻⁶ |
10⁻⁷ |
10⁻⁸ |
|
|
Green/Н5 |
19.74 ± 0.13 |
23.13 ± 0.02 |
26.76 ± 0.53 |
30.37 ± 0.44 |
33.80 ± 0.07 |
– |
|
Orange/Н7 |
21.20 ± 0.11 |
25.17 ± 0.08 |
28.23 ± 0.03 |
31.39 ± 0.39 |
35.08 ± 0.52 |
– |
|
“–” – negative result. |
||||||
Accordingly, the minimum virus amount that can be detected by the developed test system is 1.5 lg EID50/cm³ for AIV/H5 and AIV/H7.
Linear regression graphs were plotted for reactions with AIV/H5 and AIV/H7 to determine the efficiency parameters (Fig. 2). The following parameters should be taken into account for evaluation of the reaction efficiency: straight line slope (m) and correlation coefficient (R²). Ideally (at 100% efficiency), m is –3.32, but values in the range from –3.2 to –3.5 are considered optimal. Values greater than 0.98 are optimal for R² [20][22]. Determined reaction efficiency parameters for the developed test system are presented in Table 4.
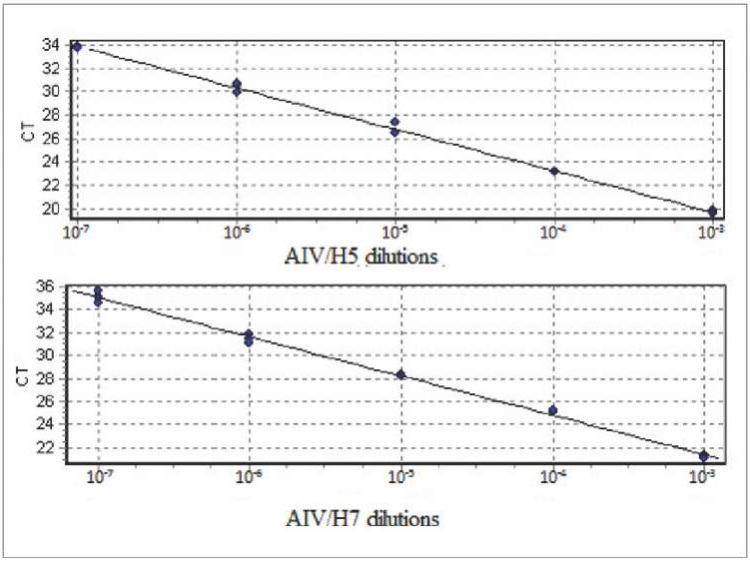
Fig. 2. Graphs of standard straight lines based on real-time RT-PCR results when 10-fold Н5 and Н7 AIV dilutions were used
Table 4
Reaction efficiency parameters for AIV/H5 and AIV/H7
|
Detection channel / AIV subtype |
Correlation coefficient (R²) |
Straight line slope (m) |
Reaction efficiency (E), % |
|
Green/Н5 |
0.997 |
–3.537 |
91.74 |
|
Orange/Н7 |
0.996 |
–3.398 |
96.92 |
The reaction efficiency for AIV/H5 (Green channel) was 91.74%, for AIV/H7 (Orange channel) was 96.92%. Parameters such as straight line slope coefficient and the coefficient of determination for AIV of both subtypes correspond to optimal values [20][22].
The reproducibility of the test system was assessed based on standard deviation (SD) for each series of 10-fold dilutions (10⁻⁷–10⁻³, n = 3). For AIV/H5 standard deviations varied from 0.02 to 0.53; for AIV/H7 standard deviations varied from 0.03 to 0.52.
For repeatability assessment, the same viruses were used at a 10⁻⁴ dilution, each sample was tested in 5 repeats. For AIV/H5, the average Ct value within the runs varied from 22.89 to 23.36; SD was 0.22–0.33; the coefficient of variation was from 0.92 to 1.17%. For AIV/H7, the average Ct value ranged from 24.53 to 25.06; the SD – from 0.18 to 0.23, and the coefficient of variation – from 0.71 to 0.94%. Repeatability values between runs for AIV/H5 were as follows: average Ct – 23.09 ± 0.32, coefficient of variation – 1.41%; for AIV/H7: average Ct – 24.53 ± 0.31, coefficient of variation – 1.25%.
A total of 434 biological samples were tested for H5 and H7 AIV RNA with the developed test system, AIV/H5 RNA was detected in 268 samples. No AIV/H7 RNA was detected in tested samples. The results obtained using the developed test system correspond to those obtained during testing of the same samples by standard molecular diagnostic methods used by the Federal Centre for Animal Health Reference Laboratory for Avian Viral Diseases [23].
CONCLUSION
The test system for detection of H5 and H7 AIV RNA by real-time RT-PCR was developed. The proposed test system parameters were determined: the specificity was 100% (AIV/H5 and AIV/H7), analytical sensitivity limit was 1.5 lg EID50/cm³ (AIV/H5 and AIV/H7), the reaction efficiency was 92% (AIV/H5) and 97% (AIV/H7). The developed test system can be used for qualitative analysis of H5 and H7 AIV RNA in biological samples from birds and other animals.
References
1. Lycett S. J., Duchatel F., Digard P. A brief history of bird flu. Philosophical Transactions of the Royal Society B: Biological Sciences. 2019; 374 (1775):20180257. https://doi.org/10.1098/rstb.2018.0257
2. Alexander D. J. A review of avian influenza in different bird species. Veterinary Microbiology. 2000; 74 (1–2): 3–13. https://doi.org/10.1016/s0378-1135(00)00160-7
3. Olsen B., Munster V. J., Wallensten A., Waldenström J., OsterhausA. D. M. E., FouchierR. A. M. GlobalpatternsofinfluenzaAvirusinwildbirds. Science. 2006; 312 (5772): 384–388. https://doi.org/10.1126/science.1122438
4. Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiological Reviews. 1992; 56 (1): 152–179. https://doi.org/10.1128/mr.56.1.152-179.1992
5. Kida Y., Okuya K., Saito T., Yamagishi J., Ohnuma A., Hattori T., et al. Structural requirements in the hemagglutinin cleavage site-coding RNA region for the generation of highly pathogenic avian influenza virus. Pathogens. 2021; 10 (12):1597. https://doi.org/10.3390/pathogens10121597
6. Spackman E. A Brief introduction to avian influenza virus. Methodsin Molecular Biology. 2020; 2123: 83–92. https://doi.org/10.1007/978-1-0716-0346-8_7
7. Pasick J., Handel K., Robinson J., Copps J., Ridd D., Hills K., et al. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. Journal of General Virology. 2005; 86 (3): 727–731. https://doi.org/10.1099/vir.0.80478-0
8. Suarez D. L., Senne D. A., Banks J., Brown I. H., Essen S. C., Lee C., et al. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerging Infectious Diseases. 2004; 10 (4): 693–699. https://doi.org/10.3201/eid1004.030396
9. Irza V. N., Volkov M. S., Varkentin A. V. O tekushchei panzootii vysokopatogennogo grippa ptits = About ongoing high pathogenicity avian influenza panzootic. Effectivnoe zhivotnovodstvo. 2022; (5): 85–86. https://elibrary.ru/rcsitl (in Russ.)
10. Liu W. J., Xiao H., Dai L., Liu D., Chen J., Qi X., et al. Avian influenza A (H7N9) virus: from low pathogenic to highly pathogenic. Frontiers of Medicine. 2021; 15 (4): 507–527. https://doi.org/10.1007/s11684-020-0814-5
11. World Organisation for Animal Health. WAHIS. Disease situation. https://wahis.woah.org/#/dashboards/country-or-disease-dashboard
12. SuttonT. C. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses. 2018; 10 (9):461. https://doi.org/10.3390/v10090461
13. O’Connell K. P., BucherJ. R., AndersonP. E., CaoC. J., KhanA. S., Gostomski M. V., Valdes J. J. Real-time fluorogenic reverse transcription-PCR assays for detection of bacteriophage MS2. Applied and Environmental Microbiology. 2006; 72 (1): 478–483. https://doi.org/10.1128/AEM.72.1.478-483.2006
14. Kalthoff D., Bogs J., Harder T., Grund C., Pohlmann A., Beer M., Hoffmann B. Nucleic acid-based detection of influenza A virussubtypes H7 and N9 with a special emphasis on the avian H7N9 virus. Eurosurveillance. 2014; 19 (10):20731. https://doi.org/10.2807/1560-7917.es2014.19.10.20731
15. Liu J., Yao L., Zhai F., Chen Y., Lei J., Bi Z., et al. Development and application of a triplex real-time PCR assay for the simultaneous detection of avian influenza virussubtype H5, H7 and H9. Journal of VirologicalMethods. 2018; 252: 49–56. https://doi.org/10.1016/j.jviromet.2017.11.005
16. Liu L., Fang B., Yu X., Li X., Lei Y. K., Chen D. Strengthened monitoring of H5 avian influenza viruses in external environment in Hubei, 2018. Current MedicalScience. 2020; 40 (1): 63–68. https://doi.org/10.1007/s11596-020-2147-7
17. Spackman E., Senne D. A., Myer T. J., Bulaga L. L., Garber L. P., Perdue M. L., et al. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of Clinical Microbiology. 2002; 40 (9): 3256–3260. https://doi.org/10.1128/jcm.40.9.3256-3260.2002
18. Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009; 55 (4): 611–622. https://doi:10.1373/clinchem.2008.112797
19. Rebrikov D. V., Samatov G. A., Trofimov D. Yu., Semenov P. A. Realtime PCR: study guide. Moscow: BINOM. Laboratoriya znanii; 2011. 225 p. https://elibrary.ru/raympl (in Russ.)
20. Green M. R., Sambrook J. Constructing a standard curve forreal-time polymerase chain reaction (PCR) experiments. Cold SpringHarbor Protocols. 2018; 2018 (10). https://doi.org/10.1101/pdb.prot095026
21. Doronin М. I., Mikhalishin D. V., Sprygin А. V., Mazloum А., Zhbanova Т. V., Gruzdev К. N., Chernyshova Е. V. Current approaches to development of real-time qPCR test-kits. Veterinary Science Today. 2023; 12 (3): 197–207. https://doi.org/10.29326/2304-196X-2023-12-3-197-207
22. Green M. R., Sambrook J. Analysis and normalization of real-time polymerase chain reaction (PCR) experimental data. Cold Spring Harbor Protocols. 2018; 2018(10). https://doi.org/10.1101/pdb.top095000
23. AndreychukD. B., AndriyasovA. V., Chvala I. A. Methodical guidelines for detection of H5 and H7 avian influenza virus RNA with real-time RT-PCR: No. 46-16. Vladimir: Federal Centre for Animal Health; 2016. 12 p. (in Russ.)
About the Authors
A. D. GrekhnevaRussian Federation
Alena D. Grekhneva, Postgraduate Student, Leading Specialist, Reference Laboratory for Avian Viral Diseases
Yur’evets, Vladimir 600901
N. G Zinyakov
Russian Federation
Nikolay G. Zinyakov, Cand. Sci. (Biology), Leading Researcher, Reference Laboratory for Avian Viral Diseases
Yur’evets, Vladimir 600901
A. V. Andriyasov
Russian Federation
Artem V. Andriyasov, Cand. Sci. (Biology), Leading Researcher, Reference Laboratory for Avian Viral Diseases
Yur’evets, Vladimir 600901
A. A. Kozlov
Russian Federation
Anton A. Kozlov, Cand. Sci. (Biology), Researcher, Reference Laboratory for Avian Viral Diseases
Yur’evets, Vladimir 600901
E. V. Ovchinnikova
Russian Federation
Evgenia V. Ovchinnikova, Cand. Sci. (Biology), Senior Researcher, Reference Laboratory for Avian Viral Diseases
Yur’evets, Vladimir 600901
D. B. Andreychuk
Russian Federation
Dmitry B. Andreychuk, Cand. Sci. (Biology), Head of Reference Laboratory for Avian Viral Diseases
Yur’evets, Vladimir 600901
P. D. Zhestkov
Russian Federation
Pavel D. Zhestkov, Leading Specialist, Reference Laboratory for Avian Viral Diseases
Yur’evets, Vladimir 600901
I. A. Chvala
Russian Federation
Ilya A. Chvala, Cand. Sci. (Veterinary Medicine), Deputy Director for Research
Yur’evets, Vladimir 600901
Review
For citations:
Grekhneva A.D., Zinyakov N.G., Andriyasov A.V., Kozlov A.A., Ovchinnikova E.V., Andreychuk D.B., Zhestkov P.D., Chvala I.A. Development of test-system for detection of H5 and H7 avian influenza virus RNA by multiplex real-time RT-PCR assay using internal control. Veterinary Science Today. 2025;14(1):40-46. https://doi.org/10.29326/2304-196X-2025-14-1-40-46
JATS XML



































