Scroll to:
Microbial species diversity and antibiotic-resistant Enterobacteriaceae spread on dairy farms
https://doi.org/10.29326/2304-196X-2025-14-3-294-301
Abstract
Introduction. Bacterial communities significantly affect the overall productivity of agricultural establishments, as animal health, milk production, and food quality and safety depend on them. Zoonotic bacteria not only have a negative impact on animal health, but also pose a risk to public health, so monitoring of the microbial species diversity on dairy farms to determine the predominant pathogen species and antibiotic resistance profiles is essential.
Objective. Study of bacterial species diversity on a dairy farm and monitoring of antibiotic resistance spread in Escherichia coli and Proteus mirabilis isolates in order to enable timely development of measures containing the spread of antibiotic-resistant microorganisms.
Materials and methods. To achieve this goal, microorganisms were identified by MALDI-ToF mass spectrometry and antibiotic susceptibility of the isolated cultures was determined using the disc diffusion test.
Results. The species diversity of microorganisms isolated from samples of cattle limb wound exudates, feces, and feed was established. Opportunistic and pathogenic Escherichia coli and Proteus mirabilis turned out to be the predominant microorganisms, and their antibiotic resistance profiles were determined. One of the Escherichia coli isolates was found to be multi-resistant; only a combination of amoxicillin and clavulanic acid proved effective in inhibiting the growth of this culture. A large proportion of Proteus mirabilis isolates were resistant to drugs included in the group of fluoroquinolones and sensitive to all other tested antibacterial agents.
Conclusion. The factors influencing the microbial species diversity in wound exudate, feces and feed were reported. Determination of Enterobacteriaceae antibiotic resistance profiles will allow for the rotation of antibacterial drugs on the studied livestock farms.
Keywords
For citations:
Zubareva V.D., Bezborodova N.A., Amineva P.G., Krivonogova A.S., Sokolova O.V., Shkuratova I.A., Isakova M.N. Microbial species diversity and antibiotic-resistant Enterobacteriaceae spread on dairy farms. Veterinary Science Today. 2025;14(3):294-301. https://doi.org/10.29326/2304-196X-2025-14-3-294-301
INTRODUCTION
Bacterial communities circulating on dairy farms significantly affect food safety, quality of dairy products, and animal health [1]. “One Health” is critically important for understanding the antibiotic resistance spread; it implies interaction between humans, animals and environment, which is especially important due to the common nature of antimicrobial-resistant bacteria in humans and animals [2]. Half a billion people in the world are engaged in animal husbandry and are directly exposed to zoonotic microorganisms [3]. Residual amounts of antimicrobials and antibiotic-resistant pathogens are often found in animal waste and pollute the soil environment and wastewater. Escherichia coli serves as a reservoir for multiple antibiotic resistance determinants, which can transmit to animals and humans through various transmission routes: direct animal contact, products of animal origin, and environmental exposure [4]. Proteus mirabilis is an opportunistic pathogenic microorganism of the Enterobacteriaceae family that causes inflammatory diseases of the skin, respiratory tract, urinary tract and gastrointestinal tract. After E. coli, it is the most common opportunistic and zoonotic bacterium found in various animals such as chickens, ducks, turtles, cattle and other domestic animals [5]. P. mirabilis is found in various environments: wastewater, soil, and gastrointestinal tract of animals and humans [6]. Failure to comply with the recommendations for prescribing antimicrobials in animal husbandry contributes to the spread of antibiotic resistance.
The relevance of this work is determined by the fact that studying the composition of bacterial communities circulating in livestock facilities will allow identifying priority microorganisms affecting animal health. The novelty of the research lies in acquiring new data on the composition of the microbiota of cattle feed, wound exudate and feces and identifying the prevalence of Enterobacteriaceae antibiotic resistance in agricultural organizations across the Sverdlovsk Oblast.
The aim of the study was to examine the species diversity of bacterial communities, as well as to monitor the spread of antibiotic-resistant enterobacteria (E. coli and P. mirabilis) on dairy farms.
MATERIALS AND METHODS
The research was conducted in 2023–2024 at the Department of Animal Genomics and Selection of Ural Federal Agrarian Scientific Research Center, Ural Branch of the Russian Academy of Sciences and at the laboratory of Quality Med LLC (Ekaterinburg). The activities were carried out in four agricultural organizations in the Sverdlovsk Oblast that are engaged in Holstein cattle breeding. A total of 61 samples were collected: exudate from the surfaces of limb wounds (25), feces (22), and feed samples (14).
Fecal samples from cows were collected with a swab probe into the tubes with modified Cary-Blair medium, which is specifically designed for intestinal pathogen transportation and their viability preservation (FecalSwabTM, Copan, Italy). Samples of the remaining biological materials were placed in tubes with Amies transport medium (ESwab®, Copan, Italy).
In the laboratory of Quality Med LLC, the quadrant streaking method was used to inoculate 10 µL of the biomaterial suspension with a sterile calibrated loop onto the following nutrient media: Columbia agar (Bio-Rad Laboratories, Inc., France) with 5% defibrinated sheep blood (E&O Laboratories Ltd., Scotland); Ploskirev agar (State Research Center for Applied Microbiology and Biotechnology, Russia); GRM-agar (State Research Center for Applied Microbiology and Biotechnology, Russia); UriSelect 4 chromogenic medium (Bio-Rad Laboratories, Inc., France); Sabouraud agar with 2% glucose and chloramphenicol (SIFIN diagnostics GmbH, Germany). The inoculated Petri dishes were transferred into the thermostat at (37 ± 1) °C and incubated for 24 hours.
The grown colonies were identified by MALDI-ToF mass spectrometry using a Vitek® MS instrument (bioMérieux, France). For this purpose, the bacterial biomass was applied to a slide spot, covered with 1 µL of matrix solution (α-cyano-3-hydroxycinnamic acid), air-dried at room temperature, and analyzed by mass spectrometry to acquire ribosomal protein spectra. The data were compared against reference databases using MYLA® software (bioMérieux, France). Genus and species identification of the isolates was performed with semi-quantitative and quantitative characterization (CFU/gram and CFU/sample).
Antibiotic susceptibility was determined according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standard method, using Mueller – Hinton agar (Bio-Rad Laboratories, Inc., France) and antibiotic discs (Bio-Rad Laboratories, Inc., France) impregnated with predefined drug concentrations (Table). The ADAGIO automatic analyzer (Bio-Rad Laboratories, Inc., France) was used to read the antibiograms. When interpreting the susceptibility categories, the EUCAST criteria were applied (E. A. Elshafiee, S. M. Nader, S. M. Dorgham, D. A. Hamza; version 12.0, effective from 01.01.2022). Despite the fact that, according to Order No. 771 of the Ministry of Agriculture of the Russian Federation of 18 November 2021, ceftazidime, cefepime, cefotaxime, ceftriaxone, and cefoperazone are prohibited for veterinary use, many of these antibacterial substances were previously included in the medicines used in the inflammatory diseases therapy in cattle, and therefore these drugs were also used to evaluate the antibiotic resistance profile of E. coli and P. mirabilis isolates.
Table
Antibacterial drugs used for assessing the antibiotic susceptibility of microorganisms
|
Drug |
Group of antibiotics |
Active substance concentration, µg |
|
Cefixime |
third-generation cephalosporins |
5 |
|
Cefpodoxime Ceftazidime |
third-generation cephalosporins |
10 |
|
Cefepime Cefotaxime Cefuroxime Ceftriaxone |
second-, third-, fourth-generation cephalosporins |
30 |
|
Cefoperazone |
third-generation cephalosporins |
75 |
|
Marbofloxacin Enrofloxacin Levofloxacin Ciprofloxacin Norfloxacin |
fluoroquinolones |
5 |
|
Gentamicin |
aminoglycosides |
10 |
|
Amoxicillin |
semi-synthetic penicillins |
30 |
|
Amoxicillin / Clavulanic acid |
semi-synthetic penicillins / beta-lactamase inhibitors |
20/10 |
RESULTS AND DISCUSSION
A total of 189 isolates were identified using MALDI-ToF method.
Among opportunistic and pathogenic microorganisms, the largest proportion in the tested wound exudate samples (n = 86) was taken by (Fig. 1): P. mirabilis (9.30%), E. coli and Trueperella pyogenes (8.14% each), Bacteroides pyogenes (6.98%), Aerococcus viridans and Prevotella melaninogenica (5.81% each), Aeromonas hydrophila and Candida catenulata (4.65% each), Bacillus altitudinis/pumilus, Mannheimia haemolytica, Streptococcus agalactiae (3.49%).
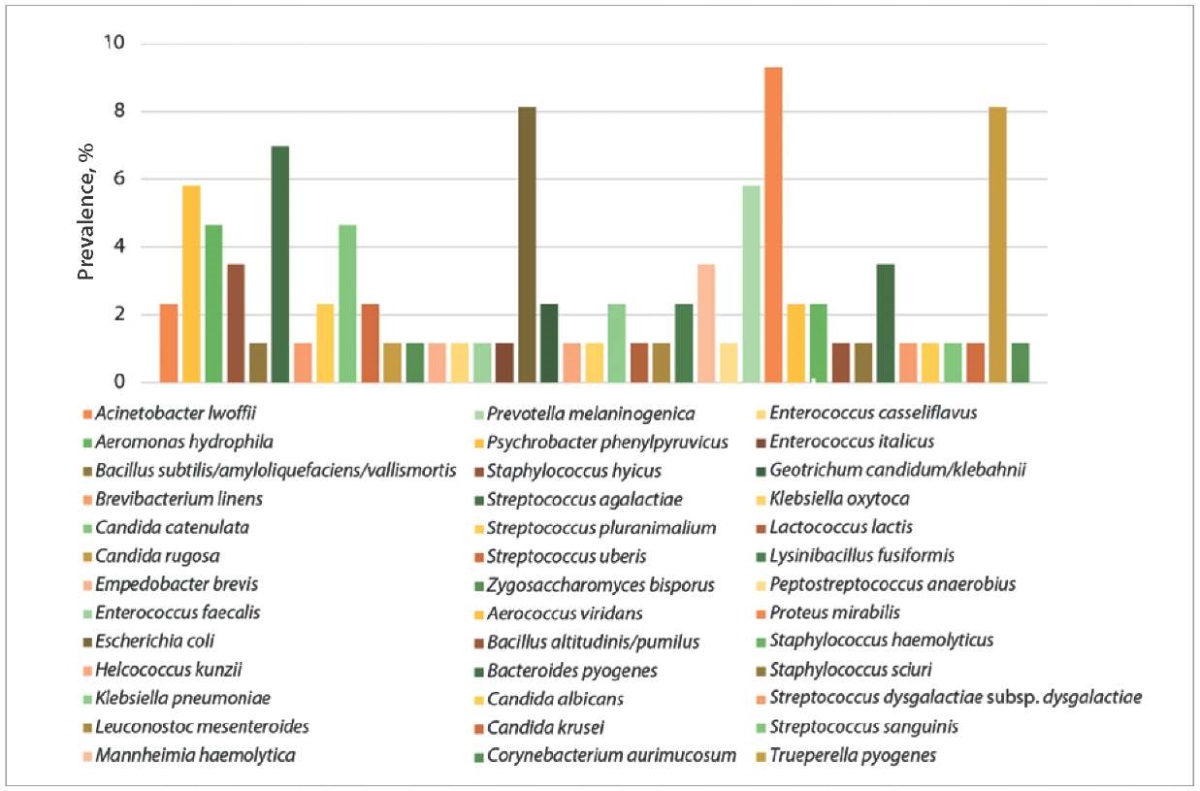
Fig. 1. Species diversity of bacterial communities isolated from wound exudates of cattle (n = 86)
According to S. C. Liegenfeld et al. [7], the following microorganisms are most often detected in the infected wound microbiota: gram-negative organisms – Pseudomonas aeruginosa, Acinetobacter baumannii, Enterobacteriaceae, E. coli, Klebsiella pneumoniae, Serratia marcescens, Enterobacter spp., Proteus spp. and Bacteroides spp.; gram-positive organisms – Staphylococcus aureus, Streptococcus spp., Enterococcus spp., Micrococcus spp., Corynebacterium spp., Streptococcus pyogenes, Corynebacterium diphtheriae and coagulase-negative staphylococci. The information on the species diversity of wound microflora published by the authors, therefore, differs from our study results.
Prevalence of P. mirabilis in animal food products and by-products is understudied [8]. P. mirabilis, as well as E. coli detections in wound exudate are most likely associated with fecal contamination of the wound surface. T. pyogenes spp. bacteria are part of the microbiota of the skin and mucous membranes of the upper respiratory tract, gastrointestinal tract, and genitourinary tract of animals and they are opportunistic microorganisms. They cause various purulent infections such as metritis, mastitis, pneumonia and abscesses leading to significant economic damage to the livestock production [9]. B. pyogenes is a representative of the oral microbiota of cats and dogs, the bites of these animals are the main risk factors of human infection. This bacterium can cause a number of inflammatory diseases, including skin and soft tissue infections, osteomyelitis, metritis, and liver abscesses [10][11]. Anaerobic bacterium P. melaninogenica, detected during the study, is involved in the footrot development and progression in cattle [12]. Representatives of the phylum Aeromonas cause diarrhea-associated diseases in piglets and pigs, colts and horses; abortions and reproductive diseases in mares, septic arthritis in colts, septicemia in dogs, mastitis in cows, polyarthritis in calves; and A. hydrophila has been identified as the only source of infection in bovine wounds [13]. S. agalactiae is a microorganism capable of inducing chronic mastitis in cows. Moreover, streptococcus of this species can colonize the gastrointestinal tract of dairy cows, and in case the wound surfaces on the limbs are contaminated with the feces, the microorganism can be detected in the wound exudate [14].
A significantly lower species diversity of opportunistic and pathogenic microorganisms was identified in bovine fecal samples (n = 59): E. coli – 28.81%, Aspergillus niger complex – 15.25%, Bacillus licheniformis – 8.47%, P. mirabilis and Enterococcus hirae – 6.78% each, other representatives of the fungal microbiota were reported (Fig. 2).
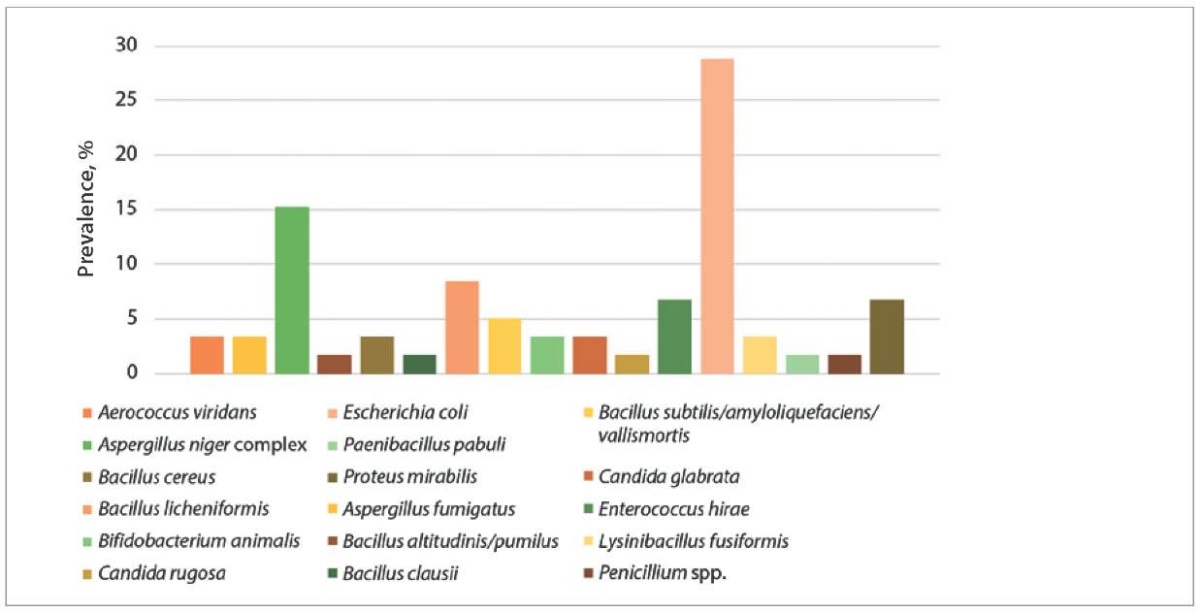
Fig. 2. Fecal microbiota species diversity in cattle (n = 59)
A decrease in the diversity of the fecal microbiota may be associated with an increase in concentrated feed in the diet [15]. A high-concentrate diet reduces the acetate-to-propionate ratio and rumen pH, adversely affecting overall health and performance of cattle [16]. Against this background, dysbiosis develops, which can lead to the increased populations of specific bacteria, such as opportunistic E. coli [16], whose predominance in the fecal microbiota of cattle was identified in our studies. Environmental contamination with fecal E. coli increases the risk of coliform mastitis in cows [17] and inflammatory diseases of the reproductive system [18].
In the feed samples (silage, haylage, and compound feed, n = 44) the predominant opportunistic and pathogenic species were the following: Bacillus altitudinis/pumilus – 9.09%, Aspergillus niger complex, Bacillus cereus, Candida krusei, Candida rugosa, and P. mirabilis – 6.82% each (Fig. 3).
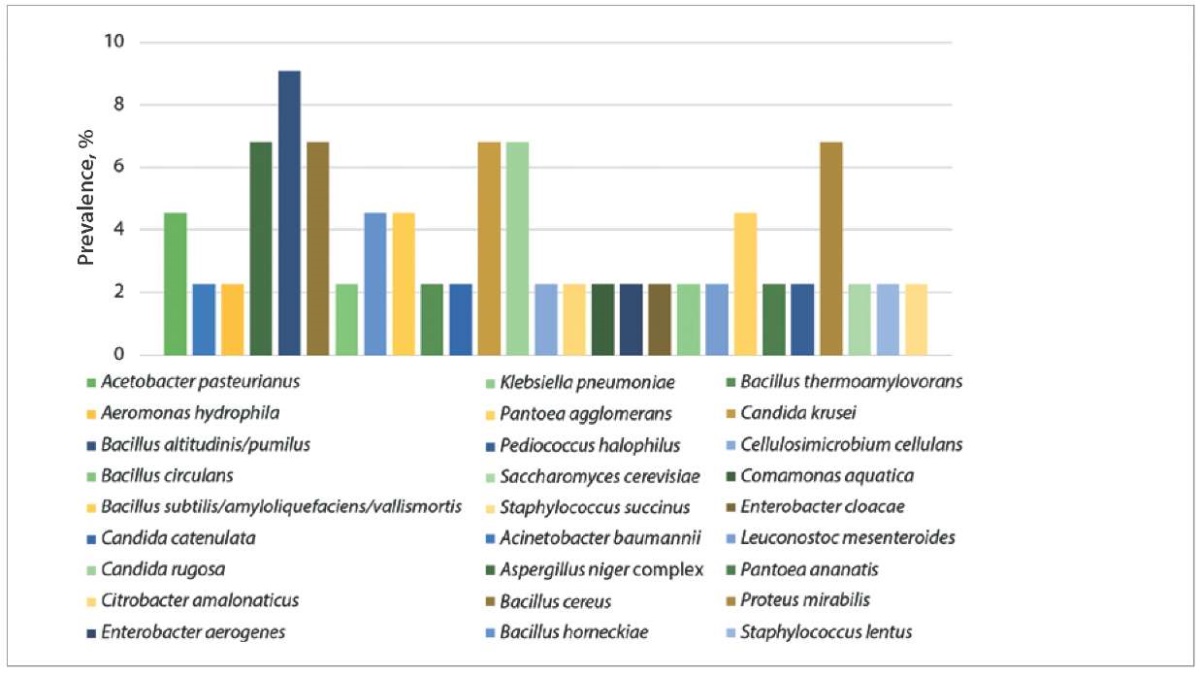
Fig. 3. Species diversity of microorganisms isolated from feed samples (n = 44)
The microorganisms typically found in silage include: Bacillus pumilus, B. licheniformis, B. coagulans, B. sphaericus, and B. cereus. Emergence of B. cereus spores in the silage is inevitable when the proper feed harvesting and storing technologies are violated. B. cereus spores pass through the gastrointestinal tract of cattle unchanged and are excreted with the feces; during milking, they can be transferred to the raw milk via contaminated nipple surfaces [19]. Presence of Candida spp., in particular C. krusei, in feed samples is an adverse indicator, since these microscopic fungi can induce mycotic mastitis in cows [20]. During our study, A. niger was detected in silage samples, which can have both positive and negative effects on cows’ health. This aspergillus species produces β-glucosidase, an enzyme that breaks down cyanogenic glycosides (toxic to cattle) thus reducing the cyanide poisoning risk and improving the silage quality [21]. At the same time, aspergillosis in cattle, especially caused by A. niger, can lead to mycotic abortions and mastitis [22]. Detection of pathogenic microorganisms in the feed samples indicates the need to develop and implement measures aimed at improving the technological processes of feed harvesting, storage and quality control in the examined livestock raising institutions.
E. coli and P. mirabilis isolates were selected to determine the antibiotic susceptibility, since these microorganisms proved to be predominant in almost all types of tested biological materials. Moreover, they are important in inducing diseases in both animals and humans.
The disk diffusion test results demonstrated that E. coli cultures (n = 17) were resistant to fluoroquinolones (35.29%) and cephalosporins (11.76%). At the same time, 80.0% of Escherichia coli isolates recovered from feces were sensitive to cefuroxime only after the increased exposure. The highest isolates’ resistance was detected to amoxicillin – 41.18%, but their resistance to amoxicillin / clavulanic acid decreased by 29.42%. Resistance to gentamicin was found in 17.65% of the isolates (Fig. 4).
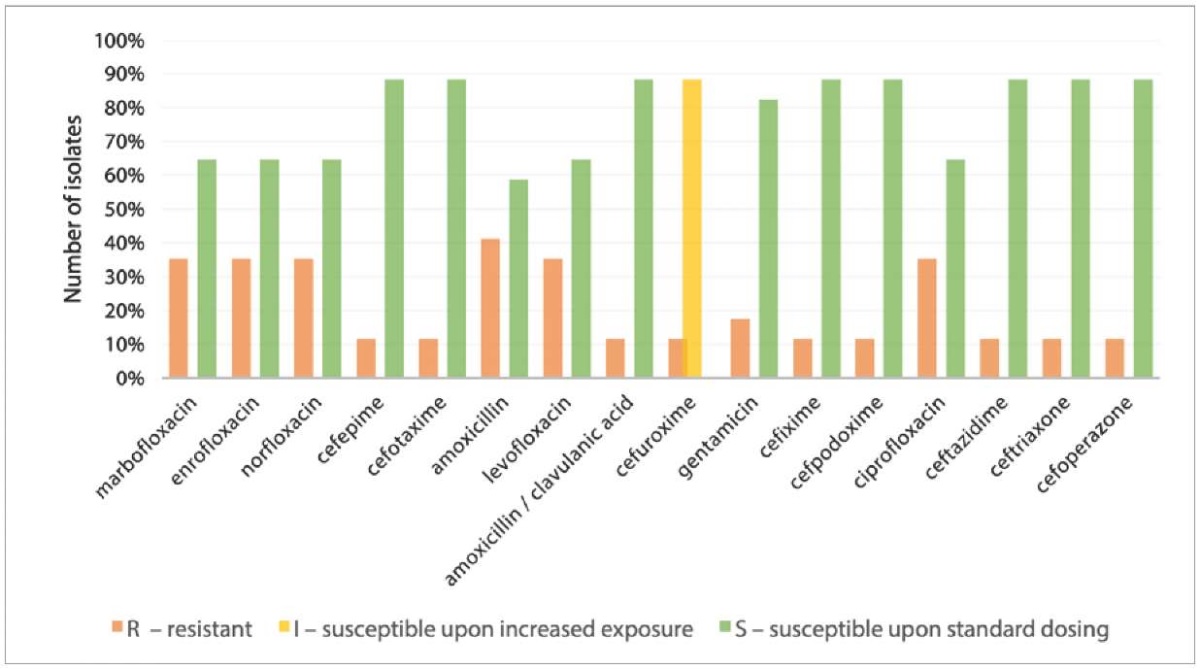
Fig. 4. Resistance of E. coli isolates to antibacterial drugs (n = 17)
The multidrug-resistant phenotype, which is defined as resistance to at least one agent from three or more antibiotic classes, was identified in one E. coli isolate. A combination of amoxicillin and clavulanic acid proved effective in suppressing this isolate’s growth. Among the multidrug-resistant pathogens widespread on dairy farms, E. coli is of particular concern, as some strains can cause foodborne infections in humans [4]. During R. Manishimwe et al. experiments [23], E. coli isolates demonstrated resistance to tetracycline (8.2%), ceftriaxone (56.8%), ciprofloxacin (77.3%) and a combination of nalidixic acid and ciprofloxacin (54.5%); that is, the frequency of resistant E. coli isolate occurrence was significantly higher than that detected in our studies.
All E. coli isolates recovered from wound exudate samples, with the exception of one resistant to amoxicillin, were found to be sensitive to all tested antibiotics. Alharbi N. S. et al. revealed that more than 50% of E. coli isolates recovered from wound discharge were resistant to cefazolin, ampicillin, cefuroxime, ciprofloxacin, mezlocillin, moxifloxacin, piperacillin, and tetracycline; 70% of the isolates produced extended-spectrum beta-lactamases [24].
Resistance to fluoroquinolones was observed in 60.0% of P. mirabilis isolates (n = 10) recovered from the wound exudate samples (Fig. 5). All isolates were susceptible to drugs of the cephalosporin group, with the exception of cefuroxime, to which 80% of isolates demonstrated susceptibility only upon the increased exposure. Resistance to amoxicillin and its combination with clavulanic acid was reported in 40% of the isolates recovered from the feces and wound exudate; 40% of isolates recovered from the wound discharge were resistant to gentamicin. In general, it is worth noting that the drugs of the cephalosporin group are effective against P. mirailis.
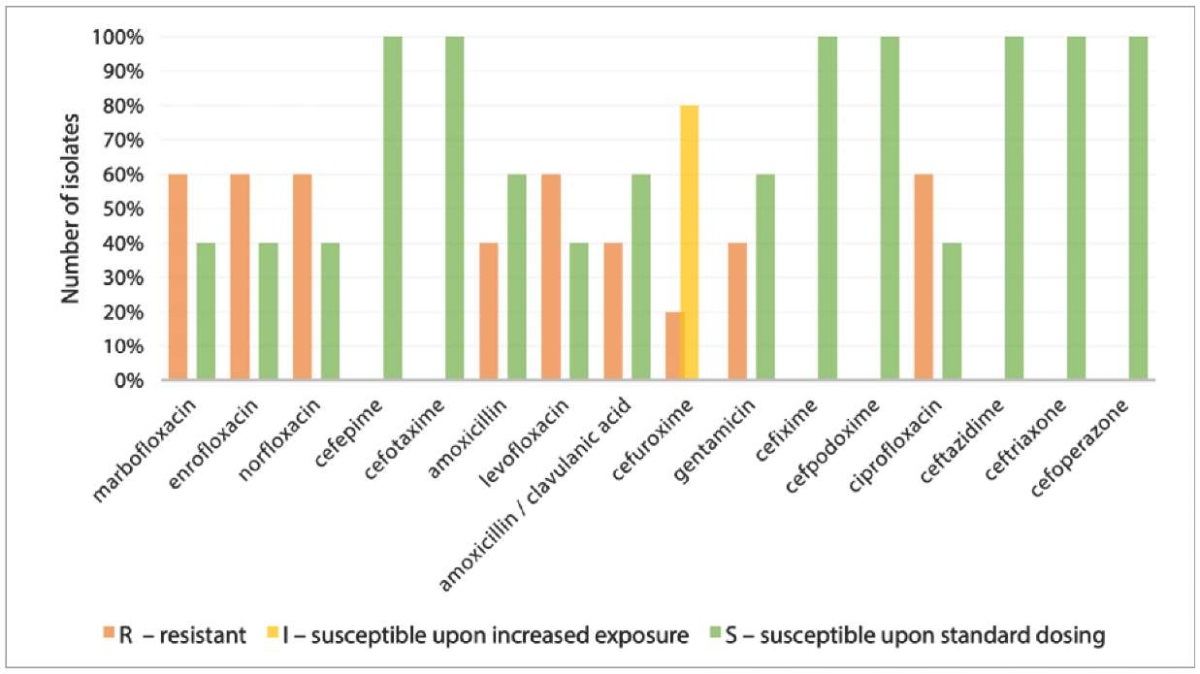
Fig. 5. Resistance of P. mirabilis isolates to antibacterial drugs (n = 10)
Previous studies have revealed that high prevalence of P. mirabilis strains resistant to penicillins, cephalosporins, and sulfonamides in chickens is a direct consequence of the use of antimicrobials in poultry farming [25][26]. The spread of antibiotic-resistant P. mirabilis in food-producing animals and in the environment of the agricultural establishments is an urgent public health problem. Transmission of P. mirabilis of multidrug resistant phenotype from animals to humans through the consumption of contaminated food or through close contact with animals has already been described [8][25].
During the studies, the species diversity of bacteria isolated from exudate samples collected from the wound surfaces on cattle limbs, feces and feed was established using the MALDI-ToF method. The data obtained can be used at the agricultural establishments to prescribe rational antibiotic therapy for both wound infections and other inflammatory diseases associated with these pathogens.
CONCLUSION
As a result of the studies, a significantly greater species diversity was revealed in the samples of wound exudate, which may mainly be associated with fecal contamination of limb wounds. The fecal microbiota was characterized by a lower species composition, which may be due to the occurrence of dysbiosis resulted from the increased proportion of the concentrated feed in the diet of cattle, while the prevalence of opportunistic E. coli (28.81%) was found in fecal samples. The predominance of pathogenic A. niger, B. cereus, and C. krusei species in feed samples indicates the need to change the technological processes of feed harvesting, storing, and quality control in the studied livestock organizations.
The antibiotic resistance profiles of E. coli and P. mirabilis were established. In E. coli cultures isolated from feces, the resistance was mainly found to drugs from the groups of fluoroquinolones (35.29%) and cephalosporins (11.76%). Notably, nearly all Escherichia isolates recovered from wound exudates demonstrated susceptibility to all tested antimicrobial agents. One E. coli isolate revealed a multidrug-resistant phenotype, and a combination of amoxicillin and clavulanic acid proved effective in suppressing its growth. It was found that almost all P. mirabilis isolates were resistant to antibacterial drugs from the group of fluoroquinolones, while the drugs from the group of cephalosporins proved effective against P. mirabilis. To prevent further growth of antimicrobial resistance, rotational use of antibacterial drugs on dairy farms should be implemented according to the identified Enterobacteriaceae resistance profiles.
Contribution of the authors: Zubareva V. D. – sample collection, literature review, text preparation, data analysis and synthesis; Bezborodova N. A. – sample collection, study design, text editing; Amineva P. G. – microbiological tests and MALDI-ToF mass spectrometry conducting, determination of antibiotic susceptibility by the disk diffusion method; Krivonogova A. S. – study concept development, paper editing; Sokolova O. V. – sample collection, administration, editing; Shkuratova I. A. – scientific support, text editing; Isakova M. N. – arrangement of the sample collection and transportation for testing.
Вклад авторов: Зубарева В. Д. – отбор проб, работа с литературой, подготовка текста, анализ и обобщение; Безбородова Н. А. – отбор проб, дизайн исследования, редактирование текста; Аминева П. Г. – проведение микробиологических исследований и MALDI-ToF масс-спектрометрии, определение антибиотикочувствительности диско-диффузионным методом; Кривоногова А. С. – составление плана исследования, редактирование текста; Соколова О. В. – отбор проб, администрирование, редактирование текста; Шкуратова И. А. – научное консультирование, редактирование текста; Исакова М. Н. – организация отбора и доставки материала для исследований.
References
1. Perdomo A., Calle A. Assessment of microbial communities in a dairy farm from a food safety perspective. International Journal of Food Microbiology. 2024; 423:110827. https://doi.org/10.1016/j.ijfoodmicro.2024.110827
2. Tyrrell C., Burgess C. M., Brennan F. P., Münzenmaier D., Drissner D., Leigh R. J., Walsh F. Genomic analysis of antimicrobial resistant Escherichiacoli isolated from manure and manured agricultural grasslands. NPJ Antimicrobials and Resistance. 2025; 3:8. https://doi.org/10.1038/s44259-025-00081-8
3. Mahmud B., Vargas R. C., Sukhum K. V., Patel S., Liao J., Hall L. R., et al. Longitudinal dynamics of farmer and livestock nasal and faecal microbiomes and resistomes. Nature Microbiology. 2024; 9: 1007–1020. https://doi.org/10.1038/s41564-024-01639-4
4. Veloo Y., Rajendiran S., Zakaria Z., Ismail R., Rahman S. A., Mansor R., Thahir S. S. A. Prevalence and antimicrobial resistance patterns of Escherichia coli in the environment, cow dung, and milk of Selangor dairy farms. Antibiotics. 2025; 14 (2):137. https://doi.org/10.3390/antibiotics14020137
5. Liu L., Dong Z., Ai S., Chen S., Dong M., Li Q., et al. Virulence-related factors and antimicrobial resistance in Proteus mirabilis isolated from domestic and stray dogs. Frontiers in Microbiology. 2023; 14:1141418. https://doi.org/10.3389/fmicb.2023.1141418
6. Al-Qurashi E., Elbnna K., Ahmad I., Abulreesh H. H. Antibiotic resistance in Proteus mirabilis: mechanism, status, and public health significance. Journal of Pure and Applied Microbiology. 2022; 16 (3): 1550–1561. https://doi.org/10.22207/JPAM.16.3.59
7. Liegenfeld S. C., Stenzel S., Rembe J.-D., Dittmer M., Ramos P., Stuermer E. K. Pathogenic and non-pathogenic microbes in the wound microbiome – how to flip the switch. Microbiology Research. 2025; 16 (2):39. https://doi.org/10.3390/microbiolres16020039
8. Chalmers G., Anderson R. E. V., Murray R., Topp E., Boerlin P. Characterization of Proteus mirabilis and associated plasmids isolated from anaerobic dairy cattle manure digesters. PloS ONE. 2023; 18 (8):e0289703. https://doi.org/10.1371/journal.pone.0289703
9. Rzewuska M., Kwiecień E., Chrobak-Chmiel D., Kizerwetter-Świda M., Stefańska I., Gieryńska M. Pathogenicity and virulence of Trueperella pyogenes: a review. International Journal of Molecular Sciences. 2019; 20 (11):2737. https://doi.org/10.3390/ijms20112737
10. Lee H. K., Walls G., Anderson G., Sullivan C., Wong C. A. Prolonged Bacteroides pyogenes infection in a patient with multiple lung abscesses. Respirology Case Reports. 2024; 12 (3):e01314. https://doi.org/10.1002/rcr2.1314
11. Cunha F., Jeon S. J., Jeong K. C., Galvão K. N. Draft genome sequences of Bacteroides pyogenes strains isolated from the uterus of Holstein dairy cows with metritis. Microbiology Resource Announcements. 2019; 8 (41):e01043-19. https://doi.org/10.1128/MRA.01043-19
12. Pyakurel S., Caddey B. J., Dias A. P., De Buck J., Morck D. W., Orsel K. Profiling bacterial communities in feedlot cattle affected with bovine foot rot and bovine digital dermatitis lesions using 16S rRNA gene sequencing and quantitative real-time PCR. BMC Microbiology. 2025; 25:158. https://doi.org/10.1186/s12866-025-03869-w
13. Awoyomi O. J., Oyewusi J. A., Talabi A. O., Oyewusi I. K., Biobaku K. T., Mustapha O. A., Agbaje M. Isolation of Aeromonas hydrophila in a case of wound infection in cattle in Nigeria. Nigerian Journal of Animal Production. 2014; 41 (1): 213–219. https://doi.org/10.51791/njap.v41i1.2726
14. Kabelitz T., Aubry E., van Vorst K., Amon T., Fulde M. The role of Streptococcus spp. in bovine mastitis. Microorganisms. 2021; 9 (7):1497. https://doi.org/10.3390/microorganisms9071497
15. Corrêa P. S., Jimenez C. R., Mendes L. W., Rymer C., Ray P., Gerdes L., et al. Taxonomy and functional diversity in the fecal microbiome of beef cattle reared in Brazilian traditional and semi-intensive production systems. Frontiers in Microbiology. 2021; 12:768480. https://doi.org/10.3389/fmicb.2021.768480
16. Auffret M. D., Dewhurst R. J., Duthie C. A., Rooke J. A., Wallace R. J., Freeman T. C., et al. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome. 2017; 5:159. https://doi.org/10.1186/s40168-017-0378-z
17. Abegewi U. A., Esemu S. N., Ndip R. N., Ndip L. M. Prevalence and risk factors of coliform-associated mastitis and antibiotic resistance of coliforms from lactating dairy cows in North West Cameroon. PloS ОNE. 2022; 17 (7):e0268247. https://doi.org/10.1371/journal.pone.0268247
18. Yamamura F., Sugiura T., Munby M., Shiokura Y., Murata R., Nakamura T., et al. Relationship between Escherichia coli virulence factors, notably kpsMTII, and symptoms of clinical metritis and endometritis in dairy cows. Journal of Veterinary Medical Science. 2022; 84 (3): 420–428. https://doi.org/10.1292/jvms.21-0586
19. Driehuis F., Wilkinson J. M., Jiang Y., Ogunade I., Adesogan A. T. Silage review: Animal and human health risks from silage. Journal of Dairy Science. 2018; 101 (5): 4093–4110. https://doi.org/10.3168/jds.2017-13836
20. Elad D., Shpigel N. Y., Winkler M., Klinger I., Fuchs V., Saran A., Faingold D. Feed contamination with Candida krusei as a probable source of mycotic mastitis in dairy cows. Journal of the American Veterinary Medical Association. 1995; 207 (5): 620–622. https://pubmed.ncbi.nlm.nih.gov/7649779
21. Zhai J., Wang B., Sun Y., Yang J., Zhou J., Wang T., et al. Effects of Aspergillus niger on cyanogenic glycosides removal and fermentation qualities of ratooning sorghum. Frontiers in Microbiology. 2023; 14:1128057. https://doi.org/10.3389/fmicb.2023.1128057
22. Seyedmousavi S., Guillot J., Arné P., de Hoog G. S., Mouton J. W., Melchers W. J. G., Verweij P. E. Aspergillus and aspergilloses in wild and domestic animals: a global health concern with parallels to human disease. Medical Mycology. 2015; 53 (8): 765–797. https://doi.org/10.1093/mmy/myv067
23. Manishimwe R., Moncada P. M., Bugarel M., Scott H. M., Loneragan G. H. Antibiotic resistance among Escherichia coli and Salmonella isolated from dairy cattle feces in Texas. PloS ONE. 2021; 16 (5):e0242390. https://doi.org/10.1371/journal.pone.0242390
24. Alharbi N. S., Khaled J. M., Kadaikunnan S., Alobaidi A. S., Sharafaddin A. H., Alyahya S. A., et al. Prevalence of Escherichia coli strains resistance to antibiotics in wound infections and raw milk. Saudi Journal of Biological Sciences. 2019; 26 (7): 1557–1562. https://doi.org/10.1016/j.sjbs.2018.11.016
25. Sarwar A., Aslam B., Mahmood S., Muzammil S., Siddique A. B., Sarwar F., et al. Distribution of multidrug-resistant Proteusmirabilis in poultry, live-stock, fish, and the related environment: One Health heed. Veterinary World. 2025; 18 (2): 446–454. https://doi.org/10.14202/vetworld.2025.446-454
26. Krivonogova A. S., Donnik I. M., Isaeva A. G., Loginov E. A., Petropavlovskiy M. V., Bespamyatnykh E. N. Antibiotic resistance of Enterobacteriaceae in microbiomes associated with poultry farming. Food Processing: Techniques and Technology. 2023; 53 (4): 710–717. https://doi.org/10.21603/2074-9414-2023-4-2472 (in Russ.)
About the Authors
V. D. ZubarevaRussian Federation
Vladlena D. Zubareva - Junior Researcher, Department of Animal Genomics and Selection, Ural Federal Agrarian Scientific Research Center, Ural Branch of the Russian Academy of Sciences.
112а Belinsky str., Ekaterinburg 620142
N. A. Bezborodova
Russian Federation
Natalia A. Bezborodova - Cand. Sci. (Veterinary Medicine), Senior Researcher, Head of Department of Animal Genomics and Selection, Ural Federal Agrarian Scientific Research Center, Ural Branch of the Russian Academy of Sciences.
112а Belinsky str., Ekaterinburg 620142
P. G. Amineva
Russian Federation
Polina G. Amineva - Microbiologist, Head of Laboratory, Quality Med LLC.
1 Mashinnaya str., Ekaterinburg 620142
A. S. Krivonogova
Russian Federation
Anna S. Krivonogova - Dr. Sci. (Biology), Leading Resercher, Department of Veterinary and Laboratory Diagnosis and Testing Laboratory, Ural Federal Agrarian Scientific Research Center, Ural Branch of the Russian Academy of Sciences.
112а Belinsky str., Ekaterinburg 620142
O. V. Sokolova
Russian Federation
Olga V. Sokolova - Dr. Sci. (Veterinary Medicine), Leading Researcher, Department of Animal Genomics and Selection, Head of Ural Scientific Research Veterinary Institute, Ural Federal Agrarian Scientific Research Center, Ural Branch of the Russian Academy of Sciences.
112а Belinsky str., Ekaterinburg 620142
I. A. Shkuratova
Russian Federation
Irina A. Shkuratova - Dr. Sci. (Veterinary Medicine), Professor, Corresponding Member of the Russian Academy of Sciences, Chief Researcher, Department of Ecology and Animals’ Noncontagious Pathology, Ural Federal Agrarian Scientific Research Center, Ural Branch of the Russian Academy of Sciences.
112а Belinsky str., Ekaterinburg 620142
M. N. Isakova
Russian Federation
Maria N. Isakova - Cand. Sci. (Veterinary Medicine), Senior Researcher, Department of Reproductive Biology and Neonatology, Ural Federal Agrarian Scientific Research Center, Ural Branch of the Russian Academy of Sciences.
112а Belinsky str., Ekaterinburg 620142
Review
For citations:
Zubareva V.D., Bezborodova N.A., Amineva P.G., Krivonogova A.S., Sokolova O.V., Shkuratova I.A., Isakova M.N. Microbial species diversity and antibiotic-resistant Enterobacteriaceae spread on dairy farms. Veterinary Science Today. 2025;14(3):294-301. https://doi.org/10.29326/2304-196X-2025-14-3-294-301



































