Scroll to:
Construction of Newcastle disease virus LaSota strain-based internal sample for rabies diagnosis with RT-PCR
https://doi.org/10.29326/2304-196X-2025-14-3-249-254
Abstract
Introduction. The following factors can impact the reliability of polymerase chain reaction diagnosis: operator errors, amplifier malfunction, presence of reaction inhibitors in the sample, poor reagent quality and others. All this can lead to the so-called false negative results.
Objective. Construction of the internal control based on heterologous Newcastle disease virus for detection of rabies virus by polymerase chain reaction.
Materials and methods. Dry live vaccine against Newcastle disease based on LaSota strain produced by the Federal Centre for Animal Health (Russia) was used as an internal control. RNA extraction from samples was performed with “Ribosorb” reagent kit (Central Research Institute for Epidemiology of the Rospotrebnadzor, Russia). Promega Corporation reagents (USA) and oligonucleotides manufactured by the Syntol Company (Russia) were used for the reverse transcription polymerase chain reaction.
Results. LaSota strain of Newcastle disease virus was selected as the target for the internal control. The primers were designed. Experiments showed that the PCR system for the internal control did not compete with the PCR system for the rabies virus when they were used together.The main parameters of reverse transcription and polymerase reaction were optimized. The developed method was validated using several key parameters: correctness, specificity, sensitivity, repeatability (intermediate precision under same conditions), and reproducibility (intermediate precision under different conditions). Validation results have shown that the method characteristics comply with the required ones.
Conclusion. Newcastle disease virus LaSota strain-based internal control has been constructed for use together with reverse transcription polymerase chain reaction assay for rabies virus detection that allows control of the assay stages in each reaction tube. This internal control after its proper optimization can be also used in experimental studies carried out at relevant research institutions for PCR diagnosis of the diseases caused by other RNA-containing viruses.
Keywords
For citations:
Chupin S.A. Construction of Newcastle disease virus LaSota strain-based internal sample for rabies diagnosis with RT-PCR. Veterinary Science Today. 2025;14(3):249-254. https://doi.org/10.29326/2304-196X-2025-14-3-249-254
INTRODUCTION
The following factors can impact the reliability of polymerase chain reaction (PCR) results: operator errors, amplifier malfunction, presence of PCR inhibitors in the sample, poor reagent quality and others [1]. This can produce false negative results, where the test reads negative despite the presence of the target agent in the sample. Use of internal controls is one of the most effective approaches aimed at making PCR assays more reliable [2]. Internal control is a nucleic acid that is added to the tested sample and undergoes all or some assay stages alongside the tested sample. In this case, specific amplification of the internal control nucleic acid fragment is observed, confirming that the entire PCR process functioned correctly.
To date, various PCR-based methods for rabies virus genome detection, using both classical PCR [3][4][5][6][7][8][9], and real-time PCR [10][11][12][13][14][15][16][17][18], have been described in the literature. However, only some of them include internal control use.
In general, internal controls are widely used in PCR diagnostics of various pathogens. There are various strategies for designing internal controls. Thus, J. Coertse et al. used artificially synthesized RNA, the sequence of which corresponded to a fragment of the rabies virus CVS strain genome [19]. Smith J. et al. used ribosomal RNA as an internal control, which, according to them, has degradation kinetics similar to viral RNA [20]. Some studies have described the internal control constructed from MS2 bacteriophage [21][22][23][24]. Genetically engineered virus-like particles were also used [25][26][27]. Plasmids containing an insert with sequences complementary to the target PCR primers are a common and effective tool for constructing internal controls [28][29][30][31]. Also, internal controls based heterologous viruses are described [32][33].
Polymerase chain reaction assay developed and described by A. E. Metlin et al. [34] having high sensitivity and specificity and providing reliable results have been used together with other methods at the Reference Laboratory for Rabies and BSE of the Federal Centre for Animal Health (Vladimir) for several years. However, modern quality standards mandate higher reliability of test results. Therefore, the study was aimed at construction of internal control system and subsequent its use in PCR diagnosis of rabies for the purpose of testing quality improvement.
MATERIALS AND METHODS
Animal brain samples submitted at the Reference Laboratory for Rabies and BSE of the Federal Centre for Animal Health (Vladimir) for testing for rabies virus were used.
ARRIAH strain of rabies virus (infectious activity in cell culture – 6.0 lg TCID50/mL) from the Federal Centre for Animal Health Collection of Microorganism Strains was used for optimization of reverse transcription polymerase chain reaction (RT-PCR) and as a positive control.
Freeze-dried live vaccine against Newcastle disease based on Newcastle disease virus LaSota strain manufactured by the Federal Centre for Animal Health (Vladimir) was used as an internal control: the vaccine filling volume – 4,000 doses, Newcastle disease virus (NDV) content – 4 × 109.7 EID50).
RNA was extracted from the samples with RIBO-sorb reagent kit (Central Research Institute of Epidemiology of the Rospotrebnadzor, Russia) according to manufacturer’s instruction.
RT-PCR was performed as described by A. E. Metlin et al. [34].
Amplicon software tool (version b08) [35] and the sequence of NDV LaSota strain (GenBank accession number – JF950510) were used for the designing primers.
Promega reagents (USA) were used for RT-PCR.
The primers were synthesized at the LLC Syntol (Russia).
PCR products were analysed by electrophoresis in 2% agarose gel with ethidium bromide staining. GelDoc gel-documenting system (Bio-Rad Laboratories, Inc., USA) was used for electrophoregram capturing.
Resulting amplicons were sequenced with the primers used for RT-PCR and BigDye Terminator Cycle Sequencing kit (Applied Biosystems, USA) on ABI Prism 3100 capillary DNA sequencer (Applied Biosystems, USA).
For validation, rabies virus RV-97 strain (obtained from the Federal Centre for Animal Health Collection of Microorganism Strains) was used as a positive standard control sample and distilled water was used as a negative standard control sample. Brain samples from various animals that tested positive for rabies virus with immunofluorescence assay (IFA) served as positive controls. Brain samples from various animals that tested negative for rabies virus with IFA served as negative control samples.
The method was assessed for its accuracy by testing the positive standard control sample in ten repeats and the negative standard control sample in ten repeats.
Ten brain samples confirmed negative for rabies virus with IFA were tested to determine the method specificity. Specificity was calculated as the percentage of true negative results to the total number of tests according to the following formula:
Sp = (TN / (TN + FP)) × 100%,
where TN – true negative result;
FP – false positive result.
Ten brain samples confirmed positive for rabies virus with IFA were tested to determine the method sensitivity. Sensitivity was calculated as the percentage of true positive results to the total number of tests according to the following formula:
Se = (TP/ (TP + FN)) × 100%,
where TP – true positive result;
FN – false negative result.
To assess the intermediate precision under same conditions (repeatability), single positive sample was analysed in triplicate under the same measurement conditions using the same equipment, operator, laboratory, and within a short time period.
To assess intermediate precision under different conditions (reproducibility), single positive sample was analysed under varying conditions. This included testing the sample in triplicate on three different days by the same operator, and in triplicate on the same day by two different operators.
RESULTS AND DISCUSSION
Heterologous viruses with similar genome structure (DNA/RNA, one/two strands) are one of the most optimal types of internal controls for detection of viruses with molecular diagnostic methods. This strategy provides end-to-end control over the assay workflow, from nucleic acid extraction through reverse transcription to PCR. In this case, the control is designed to be as similar as possible to the test sample and is processed through all analytical stages in the same reaction vessel (tube).
LaSota strain of NDV included in freeze-dried live vaccine against this disease, was selected as an internal control. The reason was that NDV, being a member of Paramyxoviridae family, has single-stranded negative-sense RNA genome similar to the rabies virus. Moreover, the vaccine can be served as a ready-to-use internal control requiring only dilution with water. This eliminates time-consuming steps such as virus cultivation and titration.
Six different pairs of primers were designed. The primers corresponded to the region of genes encoding nucleoprotein, phosphoprotein, and matrix protein, as well as to the 3'-non-coding region of NDV LaSota strain genome. The primers have been designed taking into account the annealing temperature at which the target reaction takes place (55 °C). The internal control amplicon (approximately 700 bp) was deliberately designed to be longer than the target fragment (384 bp). This ensured that the target fragment was amplified with a competitive advantage over the internal control during PCR assay [36]. The primer design was tested for its specificity using the Primer-BLAST Internet service (www.ncbi.nlm.nih.gov/tools/primer-blast).
The synthesized primer pairs were tested for their functionality with anti-ND vaccine based on NDV LaSota strain (Fig. 1).
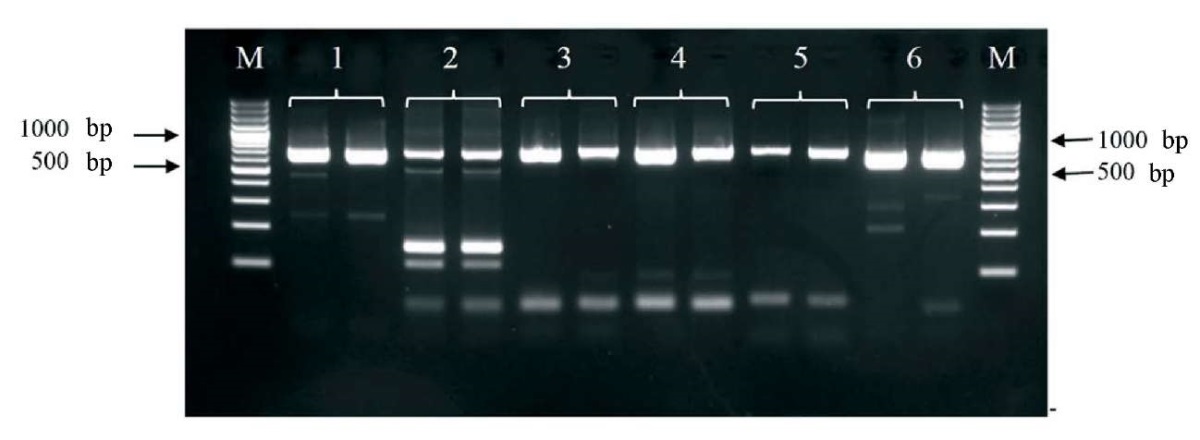
Fig. 1. Electropherogram of the PCR products using different combinations of primers for amplification Newcastle disease virus LaSota strain genome fragment. Each combination is given in duplicate. The numbers indicate the lanes with the following primer combinations: 1 – LASF815 – LASR1500; 2 – LASF1632 – LASR2337; 3 – LASF2086 – LASR2793; 4 – LASF2318 – LASR3026; 5 – LASF2598 – LASR3296; 6 – LASF107 – LASR800. М – molecular weight marker GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher Scientific) fragment length is 100 bp
Three best combinations of primers were selected based on the test results: LASF107 – LASR800, LASF2086 – LASR2793, LASF2318 – LASR3026. Each primer pair was tested in conjunction with the target PCR for rabies virus to assess potential interference between the two amplification systems. The combined use of the two systems, in all three variants, was shown to produce no undesirable effects. In addition, a ten-fold serial dilution of the vaccine was used to assess the analytical sensitivity of the internal control detection with the selected primer pairs. The pair of LASF107 – LASR800 primers showed the highest sensitivity (dilution of the initial vaccine – 1:10,000) so they were finally selected for the internal control. This pair of primers enabled amplification of a 693 nt-fragment of NDV genome. The specificity of the amplified internal control fragment was confirmed by nucleotide sequencing.
A series of experiments demonstrated that the sensitivity of the internal control detection system decreases as the concentration of rabies virus increases. In contrast, the sensitivity of the rabies virus detection system remains unchanged with increasing the internal control concentration. Thus, the internal control system does not compete with the rabies virus detection system. This is particularly important for diagnostic accuracy when the viral load in the sample is low. However, to increase the reliability of the target PCR, it was decided to use the internal control at working concentration slightly above the minimum – 1:1,000 dilution of the original vaccine. Figure 2 demonstrates the operation of both primer systems in the presence of the internal control.
Fig. 2. Electropherogram of PCR products from rabies virus-positive samples and rabies virus-negative samples. The numbers 1 and 3 indicate the lanes with rabies virus-positive samples; the numbers 2 and 4 indicate the lanes with rabies virus-negative samples. М – molecular weight marker GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher Scientific) fragments length is 100 bp
A series of experiments was performed to optimize the key parameters of reverse transcription and polymerase chain reaction. The optimal PCR parameters were found as follows: concentration of magnesium ions – 3 mM, concentration of primers – 40 mM. The internal control (10 µL of the vaccine diluted to 1:1,000 with water) was added to the test sample (50 µL of brain suspension or 100 µL of culture fluid) at the stage of RNA extraction.
Thus, the method enabling rabies virus detection in the samples with simultaneous monitoring of the assay workflow quality in each reaction tube has been developed. During the assay, known rabies virus-positive and known rabies virus-negative samples (external positive and negative controls) were also tested along with test samples. Internal control was also added to the external positive and negative controls, 10 µL per each. The presence of a 693 bp DNA fragment and the absence of a 384 bp fragment in the PCR products confirmed that the assay had been performed correctly.
Fifty-two samples confirmed positive for rabies virus with IFA and forty-eight samples confirmed negative with IFA were tested with the developed method. In all cases, the rabies virus detection results were identical to IFA results. A 693 bp fragment was always present in the PCR products when negative samples were tested, which was indicative of the internal control genome fragment amplification, i.e. the absence of a false positive result. When rabies virus-positive samples were tested, a 693 bp fragment was not always amplified. That was expected since the target reaction had a competitive advantage over the reaction with the internal control – all the resources in the reaction tube were consumed for the target fragment synthesizing. However, the 384 bp fragment was consistently detected in PCR products when the rabies virus-positive samples were tested. That was indicative of the presence of the rabies virus in the sample.
The developed method was validated using several key parameters: accuracy, specificity, sensitivity, repeatability (intermediate precision under same conditions), and reproducibility (intermediate precision under different conditions) to confirm its reliability. When the method was tested for its accuracy, the results consistent to the sample statuses (positive or negative) were obtained for all samples. Thus, the calculated accuracy of the validated method was 100%. When the method was tested for its specificity, all known rabies virus-negative samples were tested negative with the method under validation. Therefore, the calculated specificity of the method under validation was 100%. When the method was tested for its sensitivity, all positive rabies virus-positive samples were tested positive with the method under validation, so the calculated sensitivity of the method under validation was 100%. The method demonstrated good repeatability in triplicate tests. Thus, the method under validation was shown to have absolute repeatability. The method demonstrated good reproducibility in all cases. Thus, the method under validation was shown to have absolute reproducibility. Validation results showed that defined characteristics of the method comply with the required ones.
Despite the variety of internal control designs available, each has its own advantages and disadvantages. Thus, artificially synthesized RNA [19] is prone to degradation by RNases, enzymes that are typically ubiquitous in samples. The ribosomal RNA used by J. Smith et al. [20] is, in principle, more resistant to RNases due to its well-developed tertiary structures. However, the reliability and effective range of this resistance remain to be determined. MS2 bacteriophage, being a viral particle, lacks this disadvantage as it has a protein shell that protects it from RNases. However, phage cultivation is an additional stage of the work. Artificially created virus-like particles offer many advantages, including presence of protective protein shell, customizing the genome nucleotide sequence with tailored properties, and incorporation of the desired nucleic acid type. However, the creation of such particles is technologically complex and their maintaining is labour-intensive. The development of plasmids serving as internal controls in numerous domestically produced commercial diagnostic kits requires minimal economic investment, as the production processes are already well-established. However, this plasmid-based strategy limits the type of nucleic acid to DNA. When used for RNA virus diagnosis, it fails to control for critical steps such as reverse transcription. In our opinion, the use of heterologous viruses is one of the most successful strategies for designing internal controls, as it combines the advantages offered by multiple alternative approaches. Firstly, they possess a robust protein shell that provides protection against RNases. Secondly, a virus can be selected whose nucleic acid type – whether DNA or RNA, single-stranded or double-stranded-matches that of the target virus. Thirdly, these are ready-made biological objects that are relatively easily propagated in cell cultures. When using a finished product such as a vaccine (as in our case), even the cell culture cultivation and titration stages are omitted.
It should be noted that the developed internal control based on LaSota strain of NDV can also be used either directly or after optimization (e.g., if PCR parameters of the internal control system mismatch the ones of the target system) for diagnosing diseases caused by other RNA viruses.
CONCLUSION
Newcastle disease virus LaSota strain-based internal control has been constructed for use together with reverse transcription-polymerase chain reaction assay for rabies virus detection that enables control of the assay workflow in each reaction tube. This internal control system was tested for its reliability using clinical samples containing and not containing rabies virus. The constructed internal sample was successfully validated. This internal control after proper optimization can be also used in experimental studies aimed at PCR diagnosis of the diseases caused by other RNA viruses carried out at relevant research institutions.
Contribution of the author: Chupin S. A. – conceptualization; formulation and development of the key objectives and tasks; testing; analysis and interpretation of the obtained data; paper text preparation and editing.
Вклад автора: Чупин С. А. – формирование идеи, формулировка и развитие ключевых целей и задач, проведение исследований, анализ и интерпретация полученных данных, подготовка и редактирование текста.
References
1. Hoorfar J., Malorny B., Abdulmawjood A., Cook N., Wagner M., Fach P. Practical considerations in design of internal amplification controls for diagnostic PCR assays. Journal of Clinical Microbiology. 2004; 42 (5): 1863–1868. https://doi.org/10.1128/JCM.42.5.1863-1868.2004
2. Buckwalter S. P., Sloan L. M., Cunningham S. A., Espy M. J., Uhl J. R., Jones M. F., et al. Inhibition controls for qualitative real-time PCR assays: are they necessary for all specimen matrices? Journal of Clinical Microbiology. 2014; 52 (6): 2139–2143. https://doi.org/10.1128/JCM.03389-13
3. Favoretto S. R., Martorelli L. F. А., Elkhoury M. R., Zargo A. M., Durigon E. L. Rabies virus detection and phylogenetic studies in samples from an exhumed human. Clinical Infectious Diseases. 2005; 41 (3): 413–414. https://doi.org/10.1086/431766
4. Smith J. S., Orciari L. A., Yager P. A. Molecular epidemiology of rabies in the United States. Seminars in Virology. 1995; 6 (6): 387–400. https://doi.org/10.1016/S1044-5773(05)80016-2
5. De Mattos C. C., De Mattos C. A., Loza-Rubio E., Aguilar-Setién A., Orciari L. A., Smith J. S. Molecular characterization of rabies virus isolates from Mexico: implications for transmission dynamics and human risk. American Journal of Tropical Medicine and Hygiene. 1999; 61 (4): 587–597. https://doi.org/10.4269/ajtmh.1999.61.587
6. Dacheux L., Reynes J. M., Buchy P., Sivuth O., Diop B. M., Rousset D., et al. A reliable diagnosis of human rabies based on analysis of skin biopsy specimens. Clinical Infectious Diseases. 2008; 47 (11): 1410–1417. https://doi.org/10.1086/592969
7. Araújo D. B., Langoni H., Almeida M. F., Megid J. Heminested reverse-transcriptase polymerase chain reaction (hnRT-PCR) as a tool for rabies virus detection in stored and decomposed samples. BMC Research Notes. 2008; 1:17. https://doi.org/10.1186/1756-0500-1-17
8. David D., Yakobson B., Rotenberg D., Dveres N., Davidson I., Stram Y. Rabies virus detection by RT-PCR in decomposed naturally infected brains. Veterinary Microbiology. 2002; 87 (2): 111–118. https://doi.org/10.1016/s0378-1135(02)00041-x
9. Heaton P. R., Johnstone P., McElhinney L. M., Cowley R., O’Sullivan E., Whitby J. E. Heminested PCR assay for detection of six genotypes of rabies and rabies-related viruses. Journal of Clinical Microbiology. 1997; 35 (11): 2762–2766. https://doi.org/10.1128/jcm.35.11.2762-2766.1997
10. Nadin-Davis S. A., Sheen M., Wandeler A. I. Development of real-time reverse transcriptase polymerase chain reaction methods for human rabies diagnosis. Journal of Medical Virology. 2009; 81 (8): 1484–1497. https://doi.org/10.1002/jmv.21547
11. Wacharapluesadee S., Sutipanya J., Damrongwatanapokin S., Phumesin P., Chamnanpood P., Leowijuk C., Hemachudha T. Development of a TaqMan real-time RT-PCR assay for the detection of rabies virus. Journal of Virological Methods. 2008; 151 (2): 317–320. https://doi.org/10.1016/j.jviromet.2008.05.004
12. Landry M. L., Garner R., Ferguson D. Real-time nucleic acid sequence-based amplification using molecular beacons for detection of enterovirus RNA in clinical specimens. Journal of Clinical Microbiology. 2005; 43 (7): 3136–3139. https://doi.org/10.1128/JCM.43.7.3136-3139.2005
13. Hayman D. T. S., Banyard A. C., Wakeley P. R., Harkess G., Marston D., Wood J. L. N., et al. A universal real-time assay for the detection of Lyssaviruses. Journal of Virological Method. 2011; 177 (1): 87–93. https://doi.org/10.1016/j.jviromet.2011.07.002
14. Wakeley P. R., Johnson N., McElhinney L. M., Marston D., Sawyer J., Fooks A. R. Development of a real-time, TaqMan reverse transcription-PCR assay for detection and differentiation of lyssavirus genotypes 1, 5, and 6. Journal of Clinical Microbiology. 2005; 43 (6): 2786–2792. https://doi.org/10.1128/JCM.43.6.2786-2792.2005
15. Hoffmann B., Freuling C. M., Wakeley P. R., Rasmussen T. B., Leech S., Fooks A. R., et al. Improved safety for molecular diagnosis of classical rabies viruses by use of a TaqMan real-time reverse transcription-PCR“double check” strategy. Journal of Clinical Microbiology. 2010; 48 (11): 3970–3978. https://doi.org/10.1128/JCM.00612-10
16. Wadhwa A., Wilkins K., Gao J., Condori Condori R. E., Gigante C. M., Zhao H., et al. A Pan-Lyssavirus Taqman Real-Time RT-PCR Assay for the detection of highly variable Rabies virus and other lyssaviruses. PLoSNeglected Tropical Diseases. 2017; 11 (1):e0005258. https://doi.org/10.1371/journal.pntd.0005258
17. Dacheux L., Larrous F., Lavenir R., Lepelletier A., Faouzi A., Troupin C., et al. Dual combined real-time reverse transcription polymerase chain reaction assay for the diagnosis of lyssavirus infection. PLoS Neglected Tropical Diseases. 2016; 10 (7):e0004812. https://doi.org/10.1371/journal.pntd.0004812
18. Suin V., Nazé F., Francart A., Lamoral S., De Craeye S., Kalai M., Van Gucht S. A two-step lyssavirus real-time polymerase chain reaction using degenerate primers with superior sensitivity to the fluorescent antigen test. BioMed Research International. 2014; 2014:256175. https://doi.org/10.1155/2014/256175
19. Coertse J., Weyer J., Nel L. H., Markotter W. Improved PCR methods for detection of African rabies and rabies-related lyssaviruses. Journal of Clinical Microbiology. 2010; 48 (11): 3949–3955. https://doi.org/10.1128/JCM.01256-10
20. Smith J., McElhinney L. M., Heaton P. R., Black E. M., Lowings J. P. Assessment of template quality by the incorporation of an internal control into a RT-PCR for the detection of rabies and rabies-related viruses. Journal of Virological Methods. 2000; 84 (2): 107–115. https://doi.org/10.1016/s0166-0934(99)00124-x
21. Zambenedetti M. R., Pavoni D. P., Dallabona A. C., Dominguez A. C., Poersch C. O., Fragoso S. P., Krieger M. A. Internal control for real-time polymerase chain reaction based on MS2 bacteriophage for RNA viruses diagnostics. Memórias do Instituto Oswaldo Cruz. 2017; 112 (5): 339–347. https://doi.org/10.1590/0074-02760160380
22. Dreier J., Störmer M., Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. Journal of Clinical Microbiology. 2005; 43 (9): 4551–4557. https://doi.org/10.1128/JCM.43.9.4551-4557.2005
23. Felder E., Wölfel R. Development of a versatile and stable internal control system for RT-qPCR assays. Journal of Virological Methods. 2014; 208: 33–40. https://doi.org/10.1016/j.jviromet.2014.07.028
24. Blaise-Boisseau S., Hennechart-Collette C., Guillier L., Perelle S. Duplex real-time qRT-PCR for the detection of hepatitis A virus in water and raspberries using the MS2 bacteriophage as a process control. Journal of Virological Methods. 2010; 166 (1–2): 48–53. https://doi.org/10.1016/j.jviromet.2010.02.017
25. Borghetti I. A., Zambenedetti M. R., Requião L., Vieira D. S., Krieger M. A., de Cássia Pontello Rampazzo R. External control viral-like particle construction for detection of emergent arboviruses by real-time reverse-transcription PCR. BioMed Research International. 2019; 2019:2560401. https://doi.org/10.1155/2019/2560401
26. Wie Y., Yang C., Wie B., Huang J., Wang L., Meng S., et al. RNase-resistant virus-like particles containing long chimeric RNA sequences produced by two-plasmid coexpression system. Journal of Clinical Microbiology. 2008; 46 (5): 1734–1740. https://doi.org/10.1128/JCM.02248-07
27. Wang X., Liu F., Jiang L., Bao Y., Xiao Y., Wang H. Use of chimeric influenza viruses as a novel internal control for diagnostic rRT-PCR assays. Applied Microbiology and Biotechnology. 2016; 100 (4): 1667–1676. https://doi.org/10.1007/s00253-015-7042-y
28. Ursi J.-P., Ursi D., Ieven M., Pattyn S. R. Utility of an internal control for the polymerase chain reaction: application to detection of Mycoplasma pneumoniae in clinical specimens. Journal of Pathology, Microbiology and Immunology. 1992; 100 (7–12): 635–639. https://doi.org/10.1111/j.1699-0463.1992.tb03978.x
29. Zimmermann K., Mannhalter J. W. Technical aspects of quantitative competitive PCR. BioTechniques. 1996; 21 (2): 268–279. https://doi.org/10.2144/96212rv01
30. Brightwell G., Pearce M., Leslie D. Development of internal controls for PCR detection of Bacillus anthracis. Molecular and Cellular Probes. 1998; 12 (6): 367–377. https://doi.org/10.1006/mcpr.1998.0195
31. Zimmermann K., Rieger M., Groß P., Turecek P. L., Schwarz H. P. Sensitive single-stage PCR using custom-synthesized internal controls. BioTechniques. 2000; 28 (4): 694–702. https://doi.org/10.2144/00284st05
32. Roux G., Ravel C., Varlet-Marie E., Jendrowiak R., Bastien P., Sterkers Y. Inhibition of polymerase chain reaction: pathogen-specific controls are better than human gene amplification. PLoS ONE. 2019; 14 (9):e0219276. https://doi.org/10.1371/journal.pone.0219276
33. Liu J., Gratz J., Amour C., Nshama R., Walongo T., Maro A., et al. Optimization of quantitative PCR methods for enteropathogen detection. PLoS ONE. 2016; 11 (6):e0158199. https://doi.org/10.1371/journal.pone.0158199
34. Metlin A. E., Rybakov S. S., Gruzdev K. N., Neuvonen E., Cox J., Huovilainen A. Antigenic and molecular characterization of field and vaccine rabies virus strains in the Russian Federation. Developments in Biologicals. 2006; 125: 33–37. https://pubmed.ncbi.nlm.nih.gov/16878458
35. Jarman S. N. Amplicon: software for designing PCR primers on aligned DNA sequences. Bioinformatics. 2004; 20 (10): 1644–1645. https://doi.org/10.1093/bioinformatics/bth121
36. Sachadyn P., Kur J. The construction and use of a PCR internal control. Molecular and Cellular Probes. 1998; 12 (5): 259–262. https://doi.org/10.1006/mcpr.1998.0170
About the Author
S. A. ChupinRussian Federation
Sergei A. Chupin - Cand. Sci. (Biology), Leading Researcher, Reference Laboratory for Rabies and BSE, Federal Centre for Animal Health.
6 Gvardeyskaya str., Yur’evets, Vladimir 600901
Review
For citations:
Chupin S.A. Construction of Newcastle disease virus LaSota strain-based internal sample for rabies diagnosis with RT-PCR. Veterinary Science Today. 2025;14(3):249-254. https://doi.org/10.29326/2304-196X-2025-14-3-249-254



































